Targeting the AAA ATPase p97 as an Approach to Treat Cancer through Disruption of Protein Homeostasis
- PMID: 26555175
- PMCID: PMC4941640
- DOI: 10.1016/j.ccell.2015.10.002
Targeting the AAA ATPase p97 as an Approach to Treat Cancer through Disruption of Protein Homeostasis
Abstract
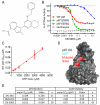
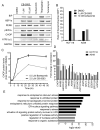
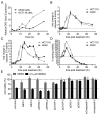
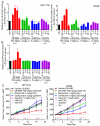
p97 is a AAA-ATPase with multiple cellular functions, one of which is critical regulation of protein homeostasis pathways. We describe the characterization of CB-5083, a potent, selective, and orally bioavailable inhibitor of p97. Treatment of tumor cells with CB-5083 leads to accumulation of poly-ubiquitinated proteins, retention of endoplasmic reticulum-associated degradation (ERAD) substrates, and generation of irresolvable proteotoxic stress, leading to activation of the apoptotic arm of the unfolded protein response. In xenograft models, CB-5083 causes modulation of key p97-related pathways, induces apoptosis, and has antitumor activity in a broad range of both hematological and solid tumor models. Molecular determinants of CB-5083 activity include expression of genes in the ERAD pathway, providing a potential strategy for patient selection.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures


Comment in
-
In Search of a Cure for Proteostasis-Addicted Cancer: A AAA Target Revealed.Cancer Cell. 2015 Nov 9;28(5):550-552. doi: 10.1016/j.ccell.2015.10.006. Cancer Cell. 2015. PMID: 26555170 Free PMC article.
References
-
- Bence NF, Sampat RM, Kopito RR. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292:1552–1555. - PubMed
-
- Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. 2001:1165–1188.
-
- Bjorkoy G, Lamark T, Johansen T. p62/SQSTM1: a missing link between protein aggregates and the autophagy machinery. Autophagy. 2006;2:138–139. - PubMed
-
- Bucheit AD, Davies MA. Emerging insights into resistance to BRAF inhibitors in melanoma. Biochemical pharmacology. 2014;87:381–389. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

