The Structural and Functional Characterization of Mammalian ADP-dependent Glucokinase
- PMID: 26555263
- PMCID: PMC4759153
- DOI: 10.1074/jbc.M115.679902
The Structural and Functional Characterization of Mammalian ADP-dependent Glucokinase
Abstract
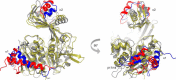
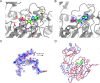
The enzyme-catalyzed phosphorylation of glucose to glucose-6-phosphate is a reaction central to the metabolism of all life. ADP-dependent glucokinase (ADPGK) catalyzes glucose-6-phosphate production, utilizing ADP as a phosphoryl donor in contrast to the more well characterized ATP-requiring hexokinases. ADPGK is found in Archaea and metazoa; in Archaea, ADPGK participates in a glycolytic role, but a function in most eukaryotic cell types remains unknown. We have determined structures of the eukaryotic ADPGK revealing a ribokinase-like tertiary fold similar to archaeal orthologues but with significant differences in some secondary structural elements. Both the unliganded and the AMP-bound ADPGK structures are in the "open" conformation. The structures reveal the presence of a disulfide bond between conserved cysteines that is positioned at the nucleotide-binding loop of eukaryotic ADPGK. The AMP-bound ADPGK structure defines the nucleotide-binding site with one of the disulfide bond cysteines coordinating the AMP with its main chain atoms, a nucleotide-binding motif that appears unique to eukaryotic ADPGKs. Key amino acids at the active site are structurally conserved between mammalian and archaeal ADPGK, and site-directed mutagenesis has confirmed residues essential for enzymatic activity. ADPGK is substrate inhibited by high glucose concentration and shows high specificity for glucose, with no activity for other sugars, as determined by NMR spectroscopy, including 2-deoxyglucose, the glucose analogue used for tumor detection by positron emission tomography.
Keywords: ADP; ADP-dependent glucokinase; AMP; NMR spectroscopy; enzyme structure; glucokinase; glucose metabolism; glucose-6-phosphate; ribokinase; x-ray crystallography.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures





References
-
- Kengen S. W., de Bok F. A., van Loo N. D., Dijkema C., Stams A. J., and de Vos W. M. (1994) Evidence for the operation of a novel Embden-Meyerhof pathway that involves ADP-dependent kinases during sugar fermentation by Pyrococcus furiosus. J. Biol. Chem. 269, 17537–17541 - PubMed
-
- Ronimus R. S., and Morgan H. W. (2004) Cloning and biochemical characterization of a novel mouse ADP-dependent glucokinase. Biochem. Biophys. Res. Commun. 315, 652–658 - PubMed
-
- Richter S., Richter J. P., Mehta S. Y., Gribble A. M., Sutherland-Smith A. J., Stowell K. M., Print C. G., Ronimus R. S., and Wilson W. R. (2012) Expression and role in glycolysis of human ADP-dependent glucokinase. Mol. Cell. Biochem. 364, 131–145 - PubMed
-
- Kamiński M. M., Sauer S. W., Kamiński M., Opp S., Ruppert T., Grigaravičius P., Grudnik P., Gröne H.-J., Krammer P. H., and Gülow K. (2012) T cell activation is driven by an ADP-dependent glucokinase linking enhanced glycolysis with mitochondrial reactive oxygen species generation. Cell Reports 2, 1300–1315 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

