Specific Inter-residue Interactions as Determinants of Human Monoacylglycerol Lipase Catalytic Competency: A ROLE FOR GLOBAL CONFORMATIONAL CHANGES
- PMID: 26555264
- PMCID: PMC4742725
- DOI: 10.1074/jbc.M115.670257
Specific Inter-residue Interactions as Determinants of Human Monoacylglycerol Lipase Catalytic Competency: A ROLE FOR GLOBAL CONFORMATIONAL CHANGES
Abstract
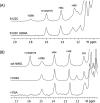
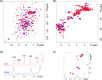
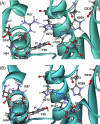
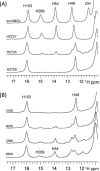
The serine hydrolase monoacylglycerol lipase (MGL) functions as the main metabolizing enzyme of 2-arachidonoyl glycerol, an endocannabinoid signaling lipid whose elevation through genetic or pharmacological MGL ablation exerts therapeutic effects in various preclinical disease models. To inform structure-based MGL inhibitor design, we report the direct NMR detection of a reversible equilibrium between active and inactive states of human MGL (hMGL) that is slow on the NMR time scale and can be modulated in a controlled manner by pH, temperature, and select point mutations. Kinetic measurements revealed that hMGL substrate turnover is rate-limited across this equilibrium. We identify a network of aromatic interactions and hydrogen bonds that regulates hMGL active-inactive state interconversion. The data highlight specific inter-residue interactions within hMGL modulating the enzymes function and implicate transitions between active (open) and inactive (closed) states of the hMGL lid domain in controlling substrate access to the enzymes active site.
Keywords: catalytic efficiency; conformational change; lipase; nuclear magnetic resonance (NMR); serine protease; spectroscopy.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures




References
-
- Makriyannis A., Mechoulam R., and Piomelli D. (2005) Therapeutic opportunities through modulation of the endocannabinoid system. Neuropharmacology 48, 1068–1071 - PubMed
-
- Bertrand T., Augé F., Houtmann J., Rak A., Vallée F., Mikol V., Berne P. F., Michot N., Cheuret D., Hoornaert C., and Mathieu M. (2010) Structural basis for human monoglyceride lipase inhibition. J. Mol. Biol. 396, 663–673 - PubMed
-
- Labar G., Bauvois C., Borel F., Ferrer J. L., Wouters J., and Lambert D. M. (2010) Crystal structure of the human monoacylglycerol lipase, a key actor in endocannabinoid signaling. Chembiochem 11, 218–227 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

