Structure of UBE2Z Enzyme Provides Functional Insight into Specificity in the FAT10 Protein Conjugation Machinery
- PMID: 26555268
- PMCID: PMC4705383
- DOI: 10.1074/jbc.M115.671545
Structure of UBE2Z Enzyme Provides Functional Insight into Specificity in the FAT10 Protein Conjugation Machinery
Abstract
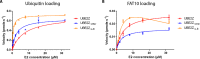
FAT10 conjugation, a post-translational modification analogous to ubiquitination, specifically requires UBA6 and UBE2Z as its activating (E1) and conjugating (E2) enzymes. Interestingly, these enzymes can also function in ubiquitination. We have determined the crystal structure of UBE2Z and report how the different domains of this E2 enzyme are organized. We further combine our structural data with mutational analyses to understand how specificity is achieved in the FAT10 conjugation pathway. We show that specificity toward UBA6 and UBE2Z lies within the C-terminal CYCI tetrapeptide in FAT10. We also demonstrate that this motif slows down transfer rates for FAT10 from UBA6 onto UBE2Z.
Keywords: FAT10; UBE2Z; UBL conjugation; crystal structure; post-translational modification (PTM); ubiquitin; ubiquitin-conjugating enzyme (E2 enzyme); ubiquitylation (ubiquitination).
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures





References
-
- Raasi S., Schmidtke G., and Groettrup M. (2001) The ubiquitin-like protein FAT10 forms covalent conjugates and induces apoptosis. J. Biol. Chem. 276, 35334–35343 - PubMed
-
- van der Veen A. G., and Ploegh H. L. (2012) Ubiquitin-like proteins. Annu. Rev. Biochem. 81, 323–357 - PubMed
-
- Pelzer C., and Groettrup M. (2010) FAT10: activated by UBA6 and functioning in protein degradation. Subcell. Biochem. 54, 238–246 - PubMed
-
- Schmidtke G., Aichem A., and Groettrup M. (2014) FAT10ylation as a signal for proteasomal degradation. Biochim. Biophys. Acta 1843, 97–102 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

