Evolutionary fate and implications of retrocopies in the African coelacanth genome
- PMID: 26555943
- PMCID: PMC4641402
- DOI: 10.1186/s12864-015-2178-9
Evolutionary fate and implications of retrocopies in the African coelacanth genome
Abstract
Background: The coelacanth is known as a "living fossil" because of its morphological resemblance to its fossil ancestors. Thus, it serves as a useful model that provides insight into the fish that first walked on land. Retrocopies are a type of novel genetic element that are likely to contribute to genome or phenotype innovations. Thus, investigating retrocopies in the coelacanth genome can determine the role of retrocopies in coelacanth genome innovations and perhaps even water-to-land adaptations.
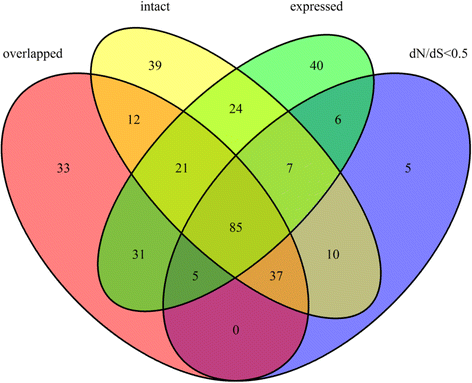
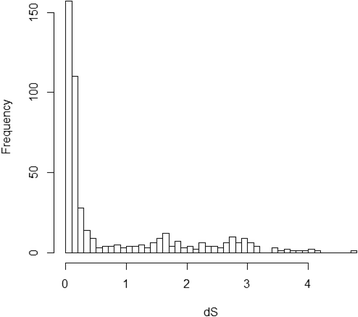
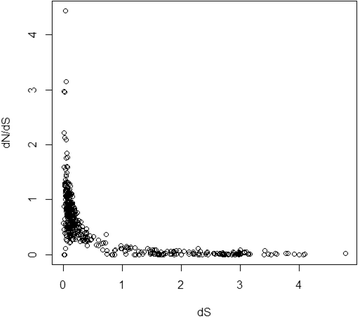
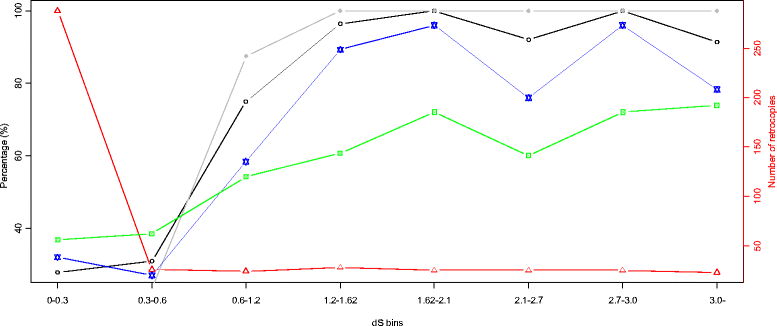
Results: We determined the dS values, dN/dS ratios, expression patterns, and enrichment of functional categories for 472 retrocopies in the African coelacanth genome. Of the retrocopies, 85-355 were shown to be potentially functional (i.e., retrogenes). The distribution of retrocopies based on their dS values revealed a burst pattern of young retrocopies in the genome. The retrocopy birth pattern was shown to be more similar to that in tetrapods than ray-finned fish, which indicates a genomic transformation that accompanied vertebrate evolution from water to land. Among these retrocopies, retrogenes were more prevalent in old than young retrocopies, which indicates that most retrocopies may have been eliminated during evolution, even though some retrocopies survived, attained biological function as retrogenes, and became old. Transcriptome data revealed that many retrocopies showed a biased expression pattern in the testis, although the expression was not specifically associated with a particular retrocopy age range. We identified 225 Ensembl genes that overlapped with the coelacanth genome retrocopies. GO enrichment analysis revealed different overrepresented GO (gene ontology) terms between these "retrocopy-overlapped genes" and the retrocopy parent genes, which indicates potential genomic functional organization produced by retrotranspositions. Among the 225 retrocopy-overlapped genes, we also identified 46 that were coelacanth-specific, which could represent a potential molecular basis for coelacanth evolution.
Conclusions: Our study identified 472 retrocopies in the coelacanth genome. Sequence analysis of these retrocopies and their parent genes, transcriptome data, and GO annotation information revealed novel insight about the potential role of genomic retrocopies in coelacanth evolution and vertebrate adaptations during the evolutionary transition from water to land.
Figures
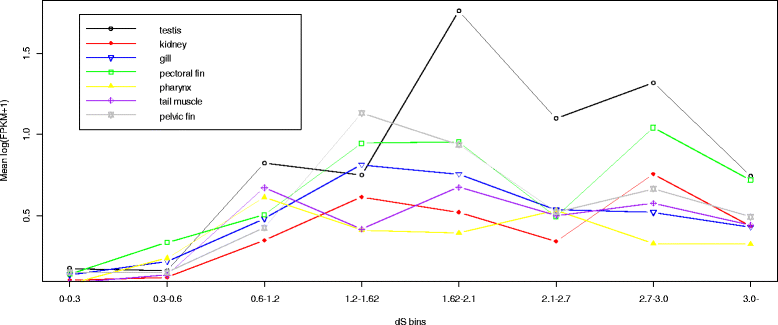
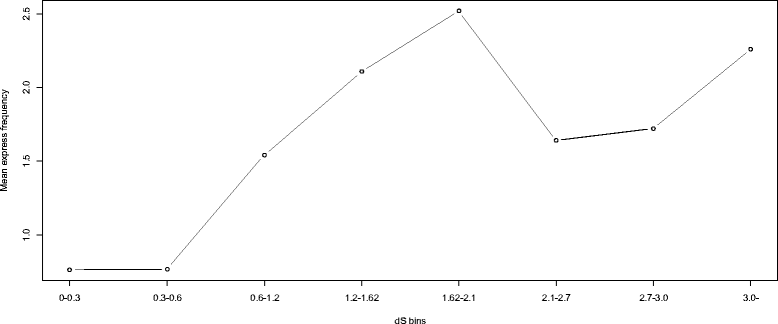
Similar articles
-
The life history of retrocopies illuminates the evolution of new mammalian genes.Genome Res. 2016 Mar;26(3):301-14. doi: 10.1101/gr.198473.115. Epub 2016 Jan 4. Genome Res. 2016. PMID: 26728716 Free PMC article.
-
A Genome-Wide Landscape of Retrocopies in Primate Genomes.Genome Biol Evol. 2015 Jul 29;7(8):2265-75. doi: 10.1093/gbe/evv142. Genome Biol Evol. 2015. PMID: 26224704 Free PMC article.
-
The African coelacanth genome provides insights into tetrapod evolution.Nature. 2013 Apr 18;496(7445):311-6. doi: 10.1038/nature12027. Nature. 2013. PMID: 23598338 Free PMC article.
-
Alternative adaptive immunity strategies: coelacanth, cod and shark immunity.Mol Immunol. 2016 Jan;69:157-69. doi: 10.1016/j.molimm.2015.09.003. Epub 2015 Sep 28. Mol Immunol. 2016. PMID: 26423359 Review.
-
Protein-Coding Genes' Retrocopies and Their Functions.Viruses. 2017 Apr 13;9(4):80. doi: 10.3390/v9040080. Viruses. 2017. PMID: 28406439 Free PMC article. Review.
Cited by
-
Comparative genomic analysis of retrogene repertoire in two green algae Volvox carteri and Chlamydomonas reinhardtii.Biol Direct. 2016 Aug 4;11:35. doi: 10.1186/s13062-016-0138-1. Biol Direct. 2016. PMID: 27487948 Free PMC article.
-
Interpopulation differences of retroduplication variations (RDVs) in rice retrogenes and their phenotypic correlations.Comput Struct Biotechnol J. 2021 Jan 5;19:600-611. doi: 10.1016/j.csbj.2020.12.046. eCollection 2021. Comput Struct Biotechnol J. 2021. PMID: 33510865 Free PMC article.
-
Identification of Retrocopies in Lepidoptera and Impact on Domestication of Silkworm.Genes (Basel). 2024 Dec 21;15(12):1641. doi: 10.3390/genes15121641. Genes (Basel). 2024. PMID: 39766908 Free PMC article.
References
-
- Robinson R. Retrocopied genes may enhance male fitness. PLoS Biol. 2005;3:e399.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

