Evolution of the EKA family of powdery mildew avirulence-effector genes from the ORF 1 of a LINE retrotransposon
- PMID: 26556056
- PMCID: PMC4641428
- DOI: 10.1186/s12864-015-2185-x
Evolution of the EKA family of powdery mildew avirulence-effector genes from the ORF 1 of a LINE retrotransposon
Abstract
Background: The Avrk1 and Avra10 avirulence (AVR) genes encode effectors that increase the pathogenicity of the fungus Blumeria graminis f.sp. hordei (Bgh), the powdery mildew pathogen, in susceptible barley plants. In resistant barley, MLK1 and MLA10 resistance proteins recognize the presence of AVRK1 and AVRA10, eliciting the hypersensitive response typical of gene for gene interactions. Avrk1 and Avra10 have more than 1350 homologues in Bgh genome, forming the EKA (Effectors homologous to Avr k 1 and Avr a 10) gene family.
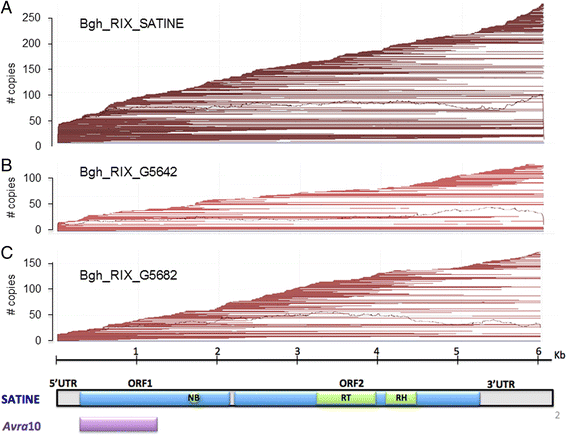
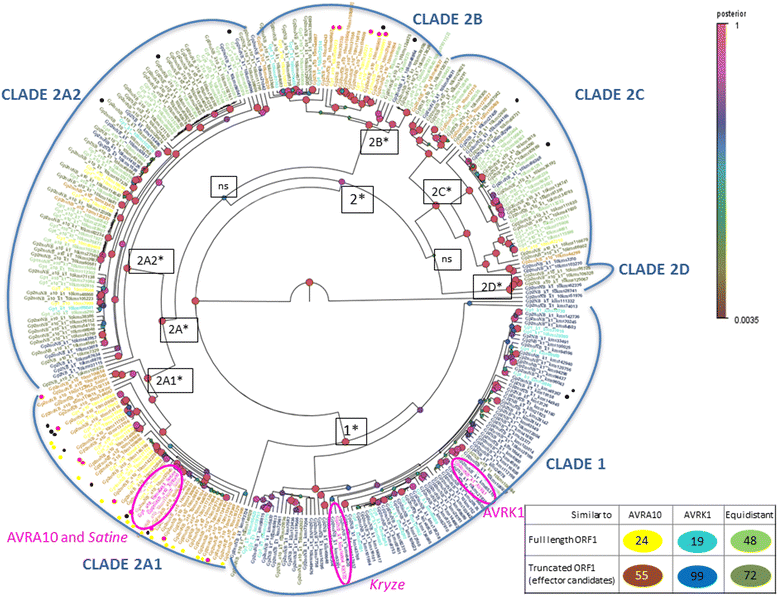
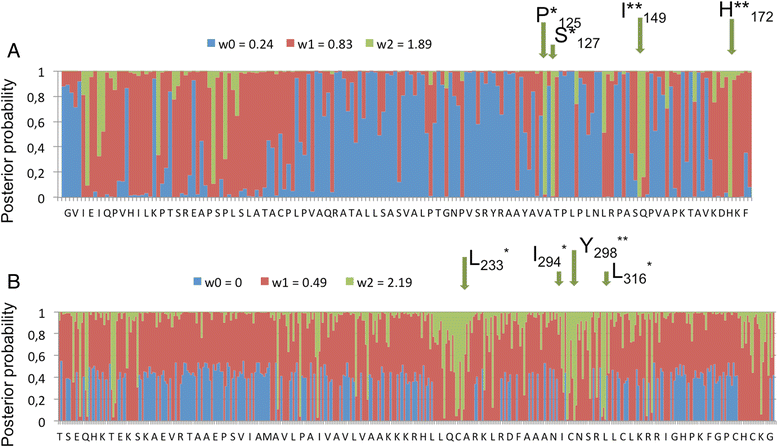
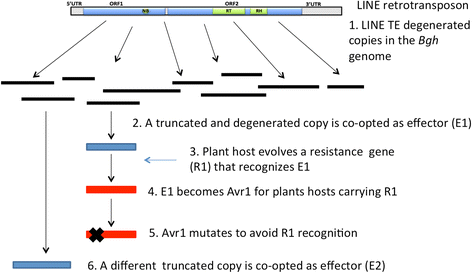
Results: We tested the hypothesis that the EKA family originated from degenerate copies of Class I LINE retrotransposons by analysing the EKA family in the genome of Bgh isolate DH14 with bioinformatic tools specially developed for the analysis of Transposable Elements (TE) in genomes. The Class I LINE retrotransposon copies homologous to Avrk1 and Avra10 represent 6.5 % of the Bgh annotated genome and, among them, we identified 293 AVR/effector candidate genes. We also experimentally identified peptides that indicated the translation of several predicted proteins from EKA family members, which had higher relative abundance in haustoria than in hyphae.
Conclusions: Our analyses indicate that Avrk1 and Avra10 have evolved from part of the ORF1 gene of Class I LINE retrotransposons. The co-option of Avra10 and Avrk1 as effectors from truncated copies of retrotransposons explains the huge number of homologues in Bgh genome that could act as dynamic reservoirs from which new effector genes may evolve. These data provide further evidence for recruitment of retrotransposons in the evolution of new biological functions.
Figures


Similar articles
-
Small RNA discovery in the interaction between barley and the powdery mildew pathogen.BMC Genomics. 2019 Jul 25;20(1):610. doi: 10.1186/s12864-019-5947-z. BMC Genomics. 2019. PMID: 31345162 Free PMC article.
-
The leucine-rich repeats in allelic barley MLA immune receptors define specificity towards sequence-unrelated powdery mildew avirulence effectors with a predicted common RNase-like fold.PLoS Pathog. 2021 Feb 3;17(2):e1009223. doi: 10.1371/journal.ppat.1009223. eCollection 2021 Feb. PLoS Pathog. 2021. PMID: 33534797 Free PMC article.
-
Multiple avirulence paralogues in cereal powdery mildew fungi may contribute to parasite fitness and defeat of plant resistance.Plant Cell. 2006 Sep;18(9):2402-14. doi: 10.1105/tpc.106.043307. Epub 2006 Aug 11. Plant Cell. 2006. PMID: 16905653 Free PMC article.
-
The role of secreted proteins in diseases of plants caused by rust, powdery mildew and smut fungi.Curr Opin Microbiol. 2007 Aug;10(4):326-31. doi: 10.1016/j.mib.2007.05.015. Epub 2007 Aug 14. Curr Opin Microbiol. 2007. PMID: 17698407 Review.
-
Specific Resistance of Barley to Powdery Mildew, Its Use and Beyond. A Concise Critical Review.Genes (Basel). 2020 Aug 21;11(9):971. doi: 10.3390/genes11090971. Genes (Basel). 2020. PMID: 32825722 Free PMC article. Review.
Cited by
-
Characterization of Two Satellite DNA Families in the Genome of the Oomycete Plant Pathogen Phytophthora parasitica.Front Genet. 2020 Jun 5;11:557. doi: 10.3389/fgene.2020.00557. eCollection 2020. Front Genet. 2020. PMID: 32582290 Free PMC article.
-
Uncovering the Mechanisms: The Role of Biotrophic Fungi in Activating or Suppressing Plant Defense Responses.J Fungi (Basel). 2024 Sep 5;10(9):635. doi: 10.3390/jof10090635. J Fungi (Basel). 2024. PMID: 39330396 Free PMC article. Review.
-
Multiple pairs of allelic MLA immune receptor-powdery mildew AVRA effectors argue for a direct recognition mechanism.Elife. 2019 Feb 19;8:e44471. doi: 10.7554/eLife.44471. Elife. 2019. PMID: 30777147 Free PMC article.
-
Avirulence Genes in Cereal Powdery Mildews: The Gene-for-Gene Hypothesis 2.0.Front Plant Sci. 2016 Mar 1;7:241. doi: 10.3389/fpls.2016.00241. eCollection 2016. Front Plant Sci. 2016. PMID: 26973683 Free PMC article.
-
Mildew-Omics: How Global Analyses Aid the Understanding of Life and Evolution of Powdery Mildews.Front Plant Sci. 2016 Feb 15;7:123. doi: 10.3389/fpls.2016.00123. eCollection 2016. Front Plant Sci. 2016. PMID: 26913042 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
- BB/C506299/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/E000983/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/J/00000605/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/J/0000A252/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

