Sorafenib, a multikinase inhibitor, induces formation of stress granules in hepatocarcinoma cells
- PMID: 26556863
- PMCID: PMC4791277
- DOI: 10.18632/oncotarget.5980
Sorafenib, a multikinase inhibitor, induces formation of stress granules in hepatocarcinoma cells
Abstract
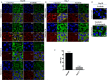
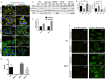
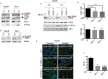
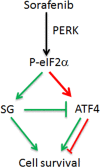
Stress granules (SGs) are cytoplasmic RNA multimeric bodies that form under stress conditions known to inhibit translation initiation. In most reported stress cases, the formation of SGs was associated with the cell recovery from stress and survival. In cells derived from cancer, SGs formation was shown to promote resistance to either proteasome inhibitors or 5-Fluorouracil used as chemotherapeutic agents. Despite these studies, the induction of SGs by chemotherapeutic drugs contributing to cancer cells resistance is still understudied. Here we identified sorafenib, a tyrosine kinase inhibitor used to treat hepatocarcinoma, as a potent chemotherapeutic inducer of SGs. The formation of SGs in sorafenib-treated hepatocarcionoma cells correlates with inhibition of translation initiation; both events requiring the phosphorylation of the translation initiation factor eIF2α. Further characterisation of the mechanism of sorafenib-induced SGs revealed PERK as the main eIF2α kinase responsible for SGs formation. Depletion experiments support the implication of PERK-eIF2α-SGs pathway in hepatocarcinoma cells resistance to sorafenib. This study also suggests the existence of an unexpected complex regulatory balance between SGs and phospho-eIF2α where SGs dampen the activation of the phospho-eIF2α-downstream ATF4 cell death pathway.
Keywords: ATF4; PERK; eIF2a; sorafenib; stress granules.
Conflict of interest statement
The authors declare no potential conflicts of interest
Figures



References
-
- Moeller B.J, Cao Y, Li C.Y, Dewhirst M.W. Radiation activates HIF-1 to regulate vascular radiosensitivity in tumors: role of reoxygenation, free radicals, and stress granules. Cancer Cell. 2004;5:429–441. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

