Sex differences in protein expression in the mouse brain and their perturbations in a model of Down syndrome
- PMID: 26557979
- PMCID: PMC4640233
- DOI: 10.1186/s13293-015-0043-9
Sex differences in protein expression in the mouse brain and their perturbations in a model of Down syndrome
Abstract
Background: While many sex differences in structure and function of the mammalian brain have been described, the molecular correlates of these differences are not broadly known. Also unknown is how sex differences at the protein level are perturbed by mutations that lead to intellectual disability (ID). Down syndrome (DS) is the most common genetic cause of ID and is due to trisomy of human chromosome 21 (Hsa21) and the resulting increased expression of Hsa21-encoded genes. The Dp(10)1Yey mouse model (Dp10) of DS is trisomic for orthologs of 39 Hsa21 protein-coding genes that map to mouse chromosome 10 (Mmu10), including four genes with known sex differences in functional properties. How these genes contribute to the DS cognitive phenotype is not known.
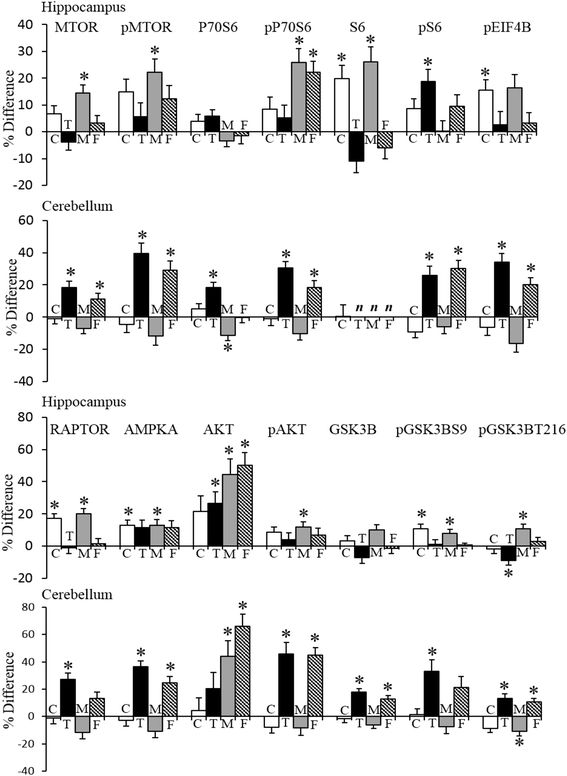
Methods: Using reverse phase protein arrays, levels of ~100 proteins/protein modifications were measured in the hippocampus, cerebellum, and cortex of female and male controls and their trisomic Dp10 littermates. Proteins were chosen for their known roles in learning/memory and synaptic plasticity and include components of the MAPK, MTOR, and apoptosis pathways, immediate early genes, and subunits of ionotropic glutamate receptors. Protein levels were compared between genotypes, sexes, and brain regions using a three-level mixed effects model and the Benjamini-Hochberg correction for multiple testing.
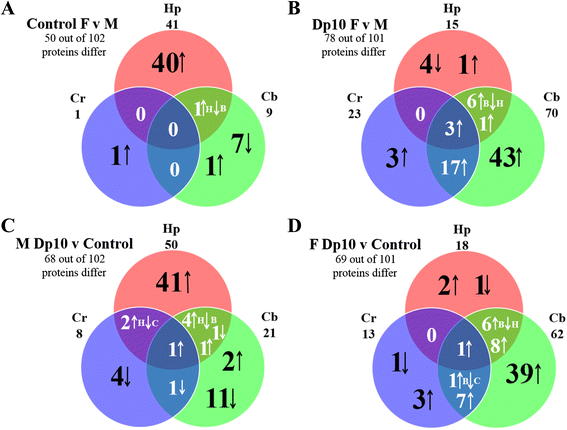
Results: In control mice, levels of approximately one half of the proteins differ significantly between females and males in at least one brain region; in the hippocampus alone, levels of 40 % of the proteins are significantly higher in females. Trisomy of the Mmu10 segment differentially affects female and male profiles, perturbing protein levels most in the cerebellum of female Dp10 and most in the hippocampus of male Dp10. Cortex is minimally affected by sex and genotype. Diverse pathways and processes are implicated in both sex and genotype differences.
Conclusions: The extensive sex differences in control mice in levels of proteins involved in learning/memory illustrate the molecular complexity underlying sex differences in normal neurological processes. The sex-specific abnormalities in the Dp10 suggest the possibility of sex-specific phenotypic features in DS and reinforce the need to use female as well as male mice, in particular in preclinical evaluations of drug responses.
Keywords: Cerebellum; Dp(10)1Yey; Hippocampus; Intellectual disability; Learning and memory deficits; Mouse chromosome 10; S100B; TRPM2; Trisomy 21.
Figures





References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

