Exome sequencing reveals a high genetic heterogeneity on familial Hirschsprung disease
- PMID: 26559152
- PMCID: PMC4642299
- DOI: 10.1038/srep16473
Exome sequencing reveals a high genetic heterogeneity on familial Hirschsprung disease
Abstract
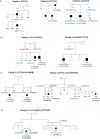
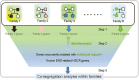
Hirschsprung disease (HSCR; OMIM 142623) is a developmental disorder characterized by aganglionosis along variable lengths of the distal gastrointestinal tract, which results in intestinal obstruction. Interactions among known HSCR genes and/or unknown disease susceptibility loci lead to variable severity of phenotype. Neither linkage nor genome-wide association studies have efficiently contributed to completely dissect the genetic pathways underlying this complex genetic disorder. We have performed whole exome sequencing of 16 HSCR patients from 8 unrelated families with SOLID platform. Variants shared by affected relatives were validated by Sanger sequencing. We searched for genes recurrently mutated across families. Only variations in the FAT3 gene were significantly enriched in five families. Within-family analysis identified compound heterozygotes for AHNAK and several genes (N = 23) with heterozygous variants that co-segregated with the phenotype. Network and pathway analyses facilitated the discovery of polygenic inheritance involving FAT3, HSCR known genes and their gene partners. Altogether, our approach has facilitated the detection of more than one damaging variant in biologically plausible genes that could jointly contribute to the phenotype. Our data may contribute to the understanding of the complex interactions that occur during enteric nervous system development and the etiopathology of familial HSCR.
Figures
References
-
- Chakravarti A, L. S. In The Metabolic and Molecular Bases of Inherited Disease, 8th ed (ed Beaudet A. R., Scriver C. R., Sly W. & Valle D.) Ch. 251, (McGraw-Hill, 2001).
-
- Alves M. M. et al. Contribution of rare and common variants determine complex diseases-Hirschsprung disease as a model. Dev Biol 382, 320–329 (2013). - PubMed
-
- Borrego S., Ruiz-Ferrer M., Fernandez R. M. & Antinolo G. Hirschsprung’s disease as a model of complex genetic etiology. Histol Histopathol 28, 1117–1136 (2013). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

