Constitutive activation of STAT3 in breast cancer cells: A review
- PMID: 26559373
- PMCID: PMC4801660
- DOI: 10.1002/ijc.29923
Constitutive activation of STAT3 in breast cancer cells: A review
Abstract
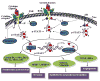
Signal transducer and activator of transcription 3 (STAT3) is constitutively activated in numerous cancer types, including more than 40% of breast cancers. In contrast to tight regulation of STAT3 as a latent transcription factor in normal cells, its signaling in breast cancer oncogenesis is multifaceted. Signaling through the IL-6/JAK/STAT3 pathway initiated by the binding of IL-6 family of cytokines (i.e., IL-6 and IL-11) to their receptors have been implicated in breast cancer development. Receptors with intrinsic kinase activity such as EGFR and VEGFR directly or indirectly induce STAT3 activation in various breast cancer types. Aberrant STAT3 signaling promotes breast tumor progression through deregulation of the expression of downstream target genes which control proliferation (Bcl-2, Bcl-xL, Survivin, Cyclin D1, c-Myc and Mcl-1), angiogenesis (Hif1α and VEGF) and epithelial-mesenchymal transition (Vimentin, TWIST, MMP-9 and MMP-7). These multiple modes of STAT3 regulation therefore make it a central linking point for a multitude of signaling processes. Extensive efforts to target STAT3 activation in breast cancer had no remarkable success in the past because the highly interconnected nature of STAT3 signaling introduces lack of selectivity in pathway identification for STAT3 targeted molecular therapies or because its role in tumorigenesis may not be as critical as it was thought. This review provides a full spectrum of STAT3's involvement in breast cancer by consolidating the knowledge about its role in breast cancer development at multiple levels: its differential regulation by different receptor signaling pathways, its downstream target genes, and modification of its transcriptional activity by its coregulatory transcription factors.
Keywords: STAT3; breast cancer; regulation; target genes; transcription factor; tumor growth.
© 2015 UICC.
Figures

References
-
- Levy DE, Darnell JE., Jr Stats: transcriptional control and biological impact. Nature reviews Molecular cell biology. 2002;3(9):651–62. - PubMed
-
- Klemm JD, Schreiber SL, Crabtree GR. Dimerization as a regulatory mechanism in signal transduction. Annual review of immunology. 1998;16:569–92. - PubMed
-
- Rane SG, Reddy EP. Janus kinases: components of multiple signaling pathways. Oncogene. 2000;19(49):5662–79. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

