Well-mixed plasma and tissue viral populations in RT-SHIV-infected macaques implies a lack of viral replication in the tissues during antiretroviral therapy
- PMID: 26559632
- PMCID: PMC4642622
- DOI: 10.1186/s12977-015-0212-2
Well-mixed plasma and tissue viral populations in RT-SHIV-infected macaques implies a lack of viral replication in the tissues during antiretroviral therapy
Abstract
Background: Determining the anatomic compartments that contribute to plasma HIV-1 is critical to understanding the sources of residual viremia during combination antiretroviral therapy (ART). We analyzed viral DNA and RNA populations in the plasma and tissues from macaques infected with SIV containing HIV-1 RT (RT-SHIV) to identify possible sources of persistent viremia and to investigate the effect of ART on viral replication in tissues. Tissues were collected at necropsy from four pigtailed macaques infected for 30 weeks with a diverse population of RT-SHIV. Two animals (6760 and 8232) were untreated and two animals (8030 and 8272) were treated with efavirenz, tenofovir, and emtricitabine for 20 weeks.
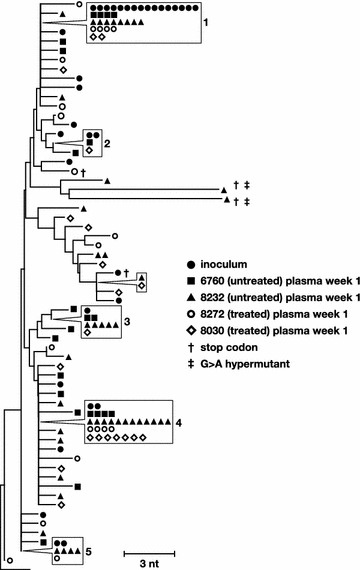
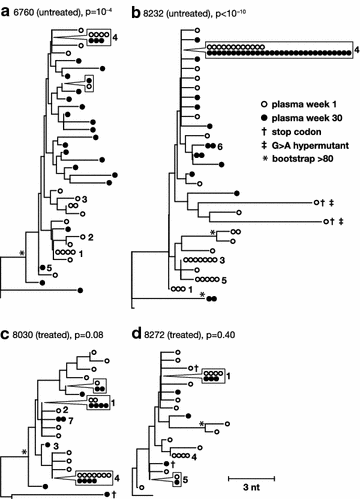
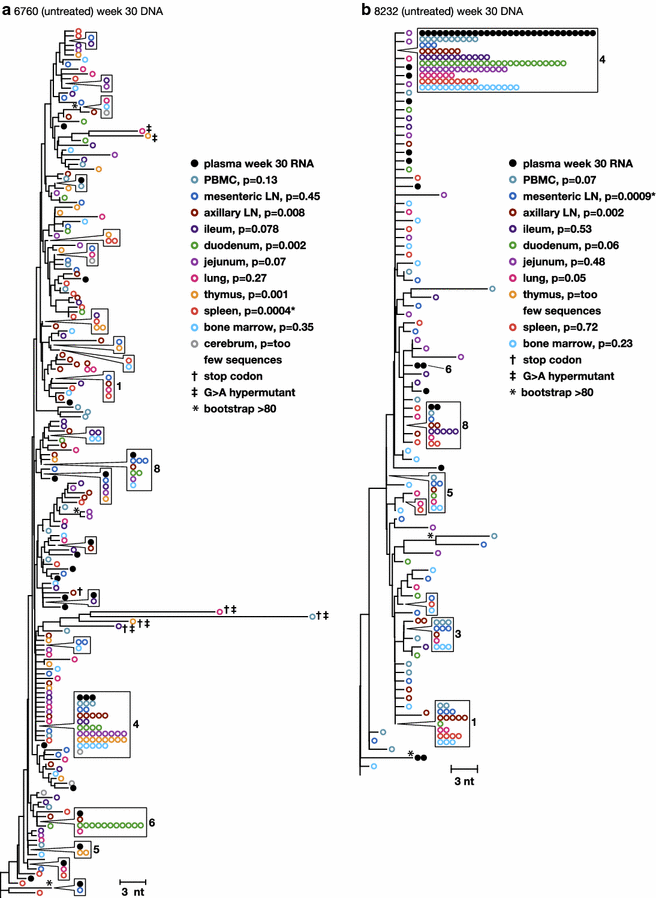
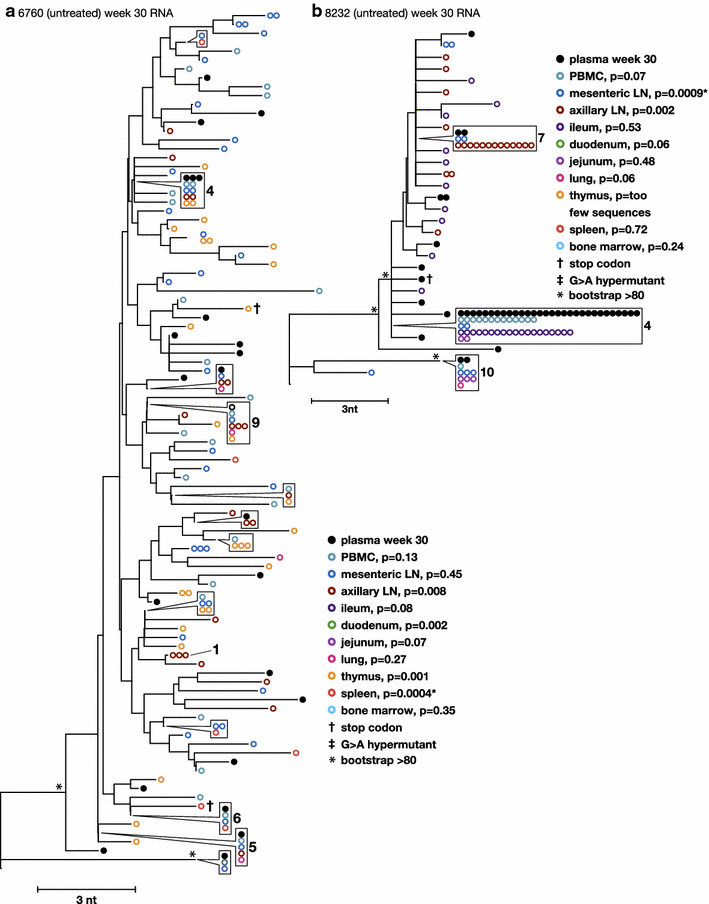
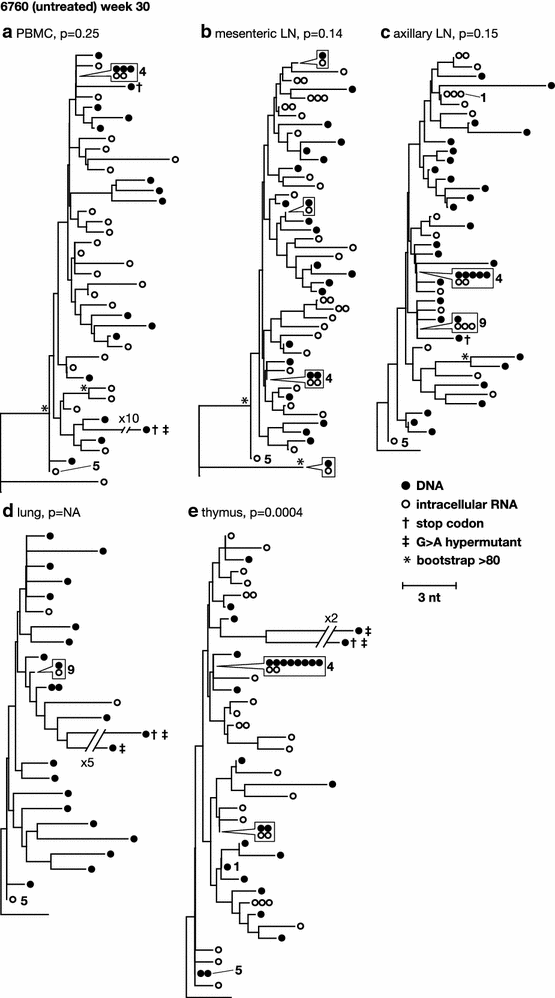
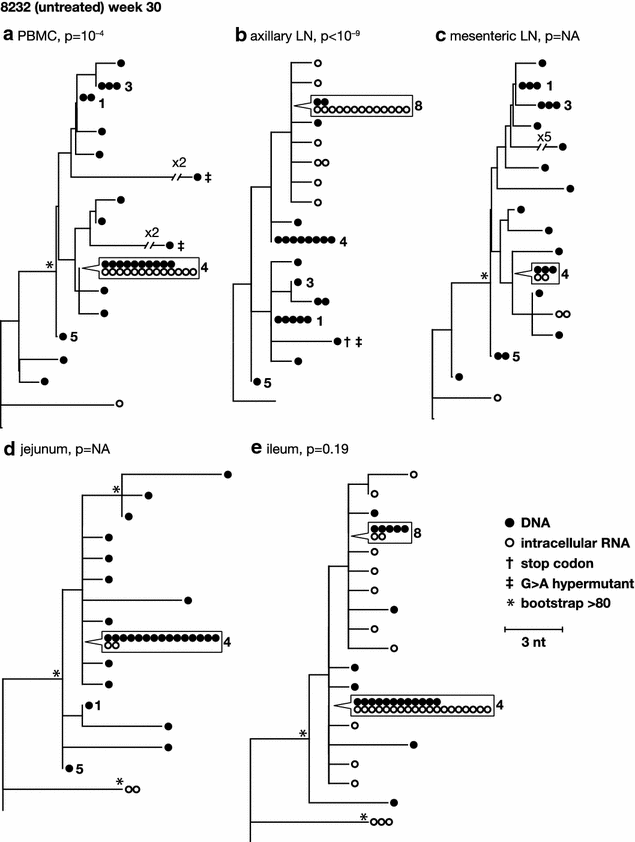
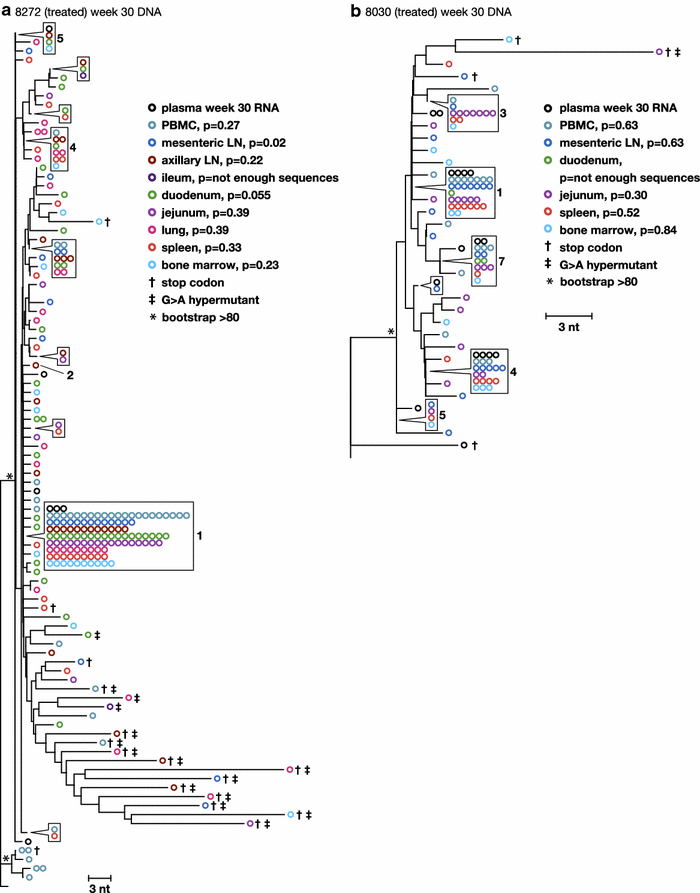
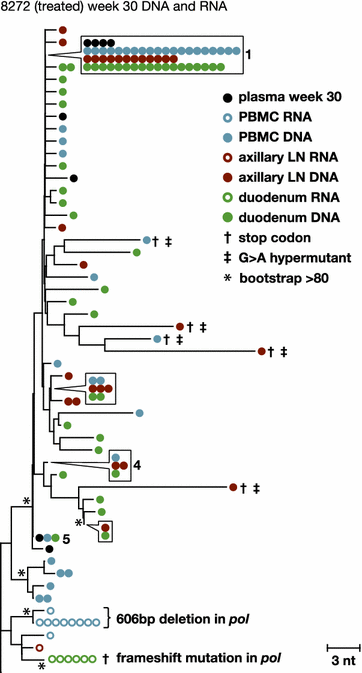
Results: A total of 1800 single-genome RT-SHIV pol and env DNA and RNA sequences were analyzed from the plasma, PBMCs, axillary and mesenteric lymph nodes, spleen, thymus, small intestine, bone marrow, lung, and brain. Analyses of intracellular DNA and RNA populations revealed that the majority of proviruses in tissues from untreated animal 8232 were not expressed, whereas a greater proportion of proviruses in tissues were expressed from 6760. Few intracellular RNA sequences were detected in treated animals and most contained inactivating mutations, such as frame shifts or large deletions. Phylogenetics showed that RT-SHIV DNA populations in tissues were not different from virus in contemporary plasma samples in the treated or untreated animals, demonstrating a lack of anatomic compartmentalization and suggesting that plasma viremia is derived from multiple tissue sources. No sequence divergence was detected in the plasma or between tissues in the treated animals after 20 weeks of ART indicating a lack of ongoing replication in tissues during treatment.
Conclusions: Virus populations in plasma and tissues did not differ significantly in either treated or untreated macaques, suggesting frequent exchange of virus or infected cells between tissues and plasma, consistent with non-compartmentalized and widely disseminated infection. There was no genetic evidence of ongoing replication in tissues during suppressive ART.
Figures
References
-
- Shiu C, Cunningham CK, Greenough T, Muresan P, Sanchez-Merino V, Carey V, Jackson JB, Ziemniak C, Belzer M, Ray SC, Luzuriaga K, Persaud D, Pediatric AIDS Clinical Trials Group P1059 Team Identification of ongoing HIV-1 replication in residual viremia during recombinant HIV-1 poxvirus immunizations in patients with clinically undetectable viral loads on durable suppressive HAART. J Virol. 2009;83:9731–9742. doi: 10.1128/JVI.00570-09. - DOI - PMC - PubMed
-
- Benito JM, Lopez M, Lozano S, Martinez P, Gonzalez-Lahoz J, Soriano V. CD38 expression on CD8 T lymphocytes as a marker of residual virus replication in chronically HIV-infected patients receiving antiretroviral therapy. AIDS Res Hum Retroviruses. 2004;20:227–233. doi: 10.1089/088922204773004950. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

