Automated amplicon design suitable for analysis of DNA variants by melting techniques
- PMID: 26559640
- PMCID: PMC4642734
- DOI: 10.1186/s13104-015-1624-8
Automated amplicon design suitable for analysis of DNA variants by melting techniques
Abstract
Background: The technological development of DNA analysis has had tremendous development in recent years, and the present deep sequencing techniques present unprecedented opportunities for detailed and high-throughput DNA variant detection. Although DNA sequencing has had an exponential decrease in cost per base pair analyzed, focused and target-specific methods are however still much in use for analysis of DNA variants. With increasing capacity in the analytical procedures, an equal demand in automated amplicon and primer design has emerged.
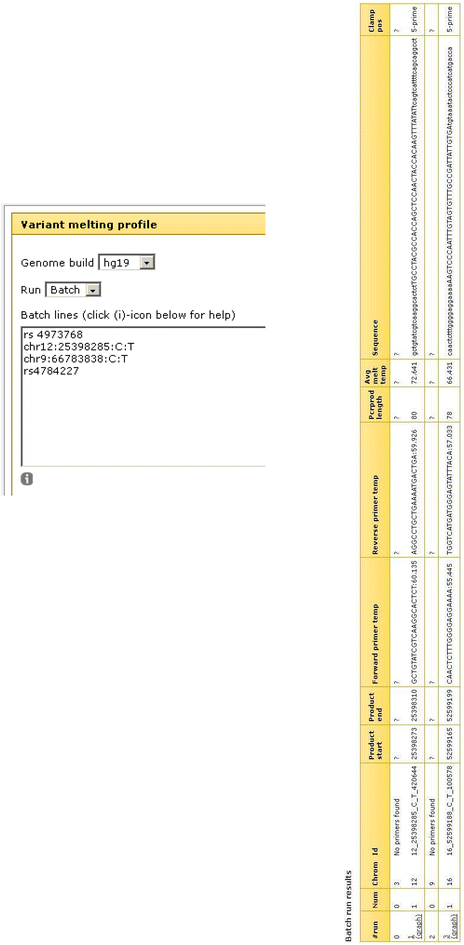
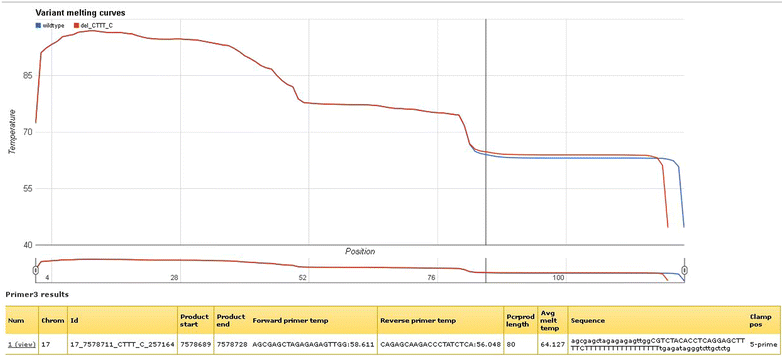
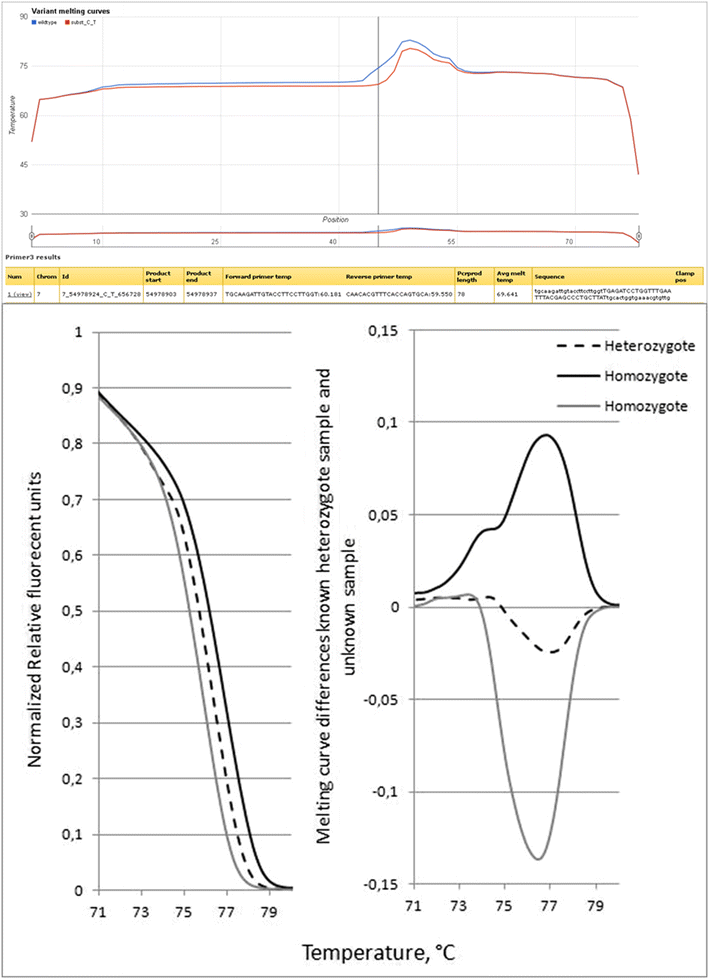
Results: We have constructed a web-based tool that is able to batch design DNA variant assay suitable for analysis by denaturing gel/capillary electrophoresis and high resolution melting. The tool is developed as a computational workflow that implements one of the most widely used primer design tools, followed by validation of primer specificity, as well as calculation and visualization of the melting properties of the resulting amplicon, with or without an artificial high melting domain attached. The tool will be useful for scientists applying DNA melting techniques in analysis of DNA variations. The tool is freely available at http://meltprimer.ous-research.no/ .
Conclusion: Herein, we demonstrate a novel tool with respect to covering the whole amplicon design workflow necessary for groups that use melting equilibrium techniques to separate DNA variants.
Figures




Similar articles
-
Simultaneous mutation scanning and genotyping by high-resolution DNA melting analysis.Nat Protoc. 2007;2(1):59-66. doi: 10.1038/nprot.2007.10. Nat Protoc. 2007. PMID: 17401339
-
Competitive amplification of differentially melting amplicons (CADMA) enables sensitive and direct detection of all mutation types by high-resolution melting analysis.Hum Mutat. 2012 Jan;33(1):264-71. doi: 10.1002/humu.21598. Epub 2011 Sep 28. Hum Mutat. 2012. PMID: 21901793
-
High throughput DNA sequence variant detection by conformation sensitive capillary electrophoresis and automated peak comparison.Genomics. 2006 Mar;87(3):427-32. doi: 10.1016/j.ygeno.2005.11.008. Epub 2006 Jan 9. Genomics. 2006. PMID: 16406726
-
High-resolution DNA melting analysis: advancements and limitations.Hum Mutat. 2009 Jun;30(6):857-9. doi: 10.1002/humu.20951. Hum Mutat. 2009. PMID: 19479960 Review.
-
DNA melting analysis.Mol Aspects Med. 2024 Jun;97:101268. doi: 10.1016/j.mam.2024.101268. Epub 2024 Mar 15. Mol Aspects Med. 2024. PMID: 38489863 Review.
Cited by
-
Applications of Probiotic-Based Multi-Components to Human, Animal and Ecosystem Health: Concepts, Methodologies, and Action Mechanisms.Microorganisms. 2022 Aug 24;10(9):1700. doi: 10.3390/microorganisms10091700. Microorganisms. 2022. PMID: 36144301 Free PMC article. Review.
-
Genetic Characterization in High-Risk Individuals from a Low-Resource City of Peru.Cancers (Basel). 2022 Nov 15;14(22):5603. doi: 10.3390/cancers14225603. Cancers (Basel). 2022. PMID: 36428697 Free PMC article.
-
Results of multigene panel testing in familial cancer cases without genetic cause demonstrated by single gene testing.Sci Rep. 2019 Dec 6;9(1):18555. doi: 10.1038/s41598-019-54517-z. Sci Rep. 2019. PMID: 31811167 Free PMC article.
-
Genetic variants of prospectively demonstrated phenocopies in BRCA1/2 kindreds.Hered Cancer Clin Pract. 2018 Jan 15;16:4. doi: 10.1186/s13053-018-0086-0. eCollection 2018. Hered Cancer Clin Pract. 2018. PMID: 29371908 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

