NPM1 histone chaperone is upregulated in glioblastoma to promote cell survival and maintain nucleolar shape
- PMID: 26559910
- PMCID: PMC4642306
- DOI: 10.1038/srep16495
NPM1 histone chaperone is upregulated in glioblastoma to promote cell survival and maintain nucleolar shape
Abstract
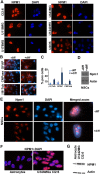
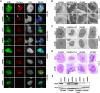
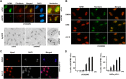
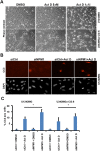
Glioblastoma (grade IV glioma) is the most common and aggressive adult brain tumor. A better understanding of the biology of glioblastoma cells is crucial to identify molecular targets stimulating cell death. NPM1 (nucleophosmin) is a multifunctional chaperone that plays an important role in cancer development. Herein, NPM1 was analyzed by immunohistochemistry in human astrocytic gliomas. NPM1 was detected in all tumors but with a significantly higher staining intensity in grade IV than in low grade tumors. Depletion of NPM1 had only modest effects on the viability of U251MG, U1242MG, and U343MGa Cl2:6 glioma cells, despite alterations in nucleolar morphology. Glioma cell cultures depleted of NPM1 exposed to micromolar levels of actinomycin D were more prone to cell death (apoptosis) compared to cultures retaining NPM1. We had previously found that NPM1 binds to linker histone H1.5. Here we could show that silencing of H1.5 triggered glioma cell apoptosis as evidenced by a marked increase in both the numbers of cleaved caspase-3(+) cells and in the amounts of cleaved PARP. Enforced expression of NPM1 suppressed apoptosis in H1.5 depleted glioma cells. Although our studies would suggest little effectiveness of targeting NPM1 alone there could be potential using it as a combination treatment.
Figures

Similar articles
-
Loss of nucleolar histone chaperone NPM1 triggers rearrangement of heterochromatin and synergizes with a deficiency in DNA methyltransferase DNMT3A to drive ribosomal DNA transcription.J Biol Chem. 2014 Dec 12;289(50):34601-19. doi: 10.1074/jbc.M114.569244. Epub 2014 Oct 27. J Biol Chem. 2014. PMID: 25349213 Free PMC article.
-
Suppression of Rac activity induces apoptosis of human glioma cells but not normal human astrocytes.Cancer Res. 2002 Apr 1;62(7):2131-40. Cancer Res. 2002. PMID: 11929835
-
Combined expressional analysis, bioinformatics and targeted proteomics identify new potential therapeutic targets in glioblastoma stem cells.Oncotarget. 2015 Sep 22;6(28):26192-215. doi: 10.18632/oncotarget.4613. Oncotarget. 2015. PMID: 26295306 Free PMC article.
-
STAT-1 expression is regulated by IGFBP-3 in malignant glioma cells and is a strong predictor of poor survival in patients with glioblastoma.J Neurosurg. 2014 Aug;121(2):374-83. doi: 10.3171/2014.4.JNS131198. Epub 2014 May 30. J Neurosurg. 2014. PMID: 24878287
-
Nucleophosmin: from structure and function to disease development.BMC Mol Biol. 2016 Aug 24;17(1):19. doi: 10.1186/s12867-016-0073-9. BMC Mol Biol. 2016. PMID: 27553022 Free PMC article. Review.
Cited by
-
The Nucleolar Protein Nucleophosmin Is Physiologically Secreted by Endothelial Cells in Response to Stress Exerting Proangiogenic Activity Both In Vitro and In Vivo.Int J Mol Sci. 2021 Apr 1;22(7):3672. doi: 10.3390/ijms22073672. Int J Mol Sci. 2021. PMID: 33916025 Free PMC article.
-
Identification of Crucial Candidate Genes and Pathways in Glioblastoma Multiform by Bioinformatics Analysis.Biomolecules. 2019 May 24;9(5):201. doi: 10.3390/biom9050201. Biomolecules. 2019. PMID: 31137733 Free PMC article.
-
Heat Shock Proteins in Glioblastoma Biology: Where Do We Stand?Int J Mol Sci. 2019 Nov 18;20(22):5794. doi: 10.3390/ijms20225794. Int J Mol Sci. 2019. PMID: 31752169 Free PMC article. Review.
-
The Targeting of RNA Polymerase I Transcription Using CX-5461 in Combination with Radiation Enhances Tumour Cell Killing Effects in Human Solid Cancers.Cancers (Basel). 2019 Sep 25;11(10):1429. doi: 10.3390/cancers11101429. Cancers (Basel). 2019. PMID: 31557908 Free PMC article.
-
Nucleophosmin in Its Interaction with Ligands.Int J Mol Sci. 2020 Jul 10;21(14):4885. doi: 10.3390/ijms21144885. Int J Mol Sci. 2020. PMID: 32664415 Free PMC article. Review.
References
-
- Furnari F. B. et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 21, 2683–2710 (2007). - PubMed
-
- Belkacemi L. & Hebb M. O. HSP27 knockdown produces synergistic induction of apoptosis by HSP90 and kinase inhibitors in glioblastoma multiforme. Anticancer Res. 34, 4915–4927 (2014). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

