The Structure of the Cyprinid herpesvirus 3 ORF112-Zα·Z-DNA Complex Reveals a Mechanism of Nucleic Acids Recognition Conserved with E3L, a Poxvirus Inhibitor of Interferon Response
- PMID: 26559969
- PMCID: PMC4692202
- DOI: 10.1074/jbc.M115.679407
The Structure of the Cyprinid herpesvirus 3 ORF112-Zα·Z-DNA Complex Reveals a Mechanism of Nucleic Acids Recognition Conserved with E3L, a Poxvirus Inhibitor of Interferon Response
Abstract
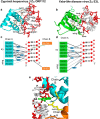
In vertebrate species, the innate immune system down-regulates protein translation in response to viral infection through the action of the double-stranded RNA (dsRNA)-activated protein kinase (PKR). In some teleost species another protein kinase, Z-DNA-dependent protein kinase (PKZ), plays a similar role but instead of dsRNA binding domains, PKZ has Zα domains. These domains recognize the left-handed conformer of dsDNA and dsRNA known as Z-DNA/Z-RNA. Cyprinid herpesvirus 3 infects common and koi carp, which have PKZ, and encodes the ORF112 protein that itself bears a Zα domain, a putative competitive inhibitor of PKZ. Here we present the crystal structure of ORF112-Zα in complex with an 18-bp CpG DNA repeat, at 1.5 Å. We demonstrate that the bound DNA is in the left-handed conformation and identify key interactions for the specificity of ORF112. Localization of ORF112 protein in stress granules induced in Cyprinid herpesvirus 3-infected fish cells suggests a functional behavior similar to that of Zα domains of the interferon-regulated, nucleic acid surveillance proteins ADAR1 and DAI.
Keywords: DNA viruses; X-ray crystallography; Zalpha domain; interferon; protein-nucleic acid interaction; stress granule.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures






References
-
- Wang A. H., Quigley G. J., Kolpak F. J., Crawford J. L., van Boom J. H., van der Marel G., and Rich A. (1979) Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature 282, 680–686 - PubMed
-
- Rich A., and Zhang S. (2003) Timeline: Z-DNA: the long road to biological function. Nat. Rev. Genet. 4, 566–572 - PubMed
-
- Athanasiadis A. (2012) Zalpha-domains: at the intersection between RNA editing and innate immunity. Semin. Cell Dev. Biol. 23, 275–280 - PubMed
-
- Schwartz T., Rould M. A., Lowenhaupt K., Herbert A., and Rich A. (1999) Crystal structure of the Zalpha domain of the human editing enzyme ADAR1 bound to left-handed Z-DNA. Science 284, 1841–1845 - PubMed
-
- Schwartz T., Behlke J., Lowenhaupt K., Heinemann U., and Rich A. (2001) Structure of the DLM-1-Z-DNA complex reveals a conserved family of Z-DNA-binding proteins. Nat. Struct. Biol. 8, 761–765 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Research Materials

