Adaptive Evolution of Eel Fluorescent Proteins from Fatty Acid Binding Proteins Produces Bright Fluorescence in the Marine Environment
- PMID: 26561348
- PMCID: PMC4641735
- DOI: 10.1371/journal.pone.0140972
Adaptive Evolution of Eel Fluorescent Proteins from Fatty Acid Binding Proteins Produces Bright Fluorescence in the Marine Environment
Abstract
We report the identification and characterization of two new members of a family of bilirubin-inducible fluorescent proteins (FPs) from marine chlopsid eels and demonstrate a key region of the sequence that serves as an evolutionary switch from non-fluorescent to fluorescent fatty acid-binding proteins (FABPs). Using transcriptomic analysis of two species of brightly fluorescent Kaupichthys eels (Kaupichthys hyoproroides and Kaupichthys n. sp.), two new FPs were identified, cloned and characterized (Chlopsid FP I and Chlopsid FP II). We then performed phylogenetic analysis on 210 FABPs, spanning 16 vertebrate orders, and including 163 vertebrate taxa. We show that the fluorescent FPs diverged as a protein family and are the sister group to brain FABPs. Our results indicate that the evolution of this family involved at least three gene duplication events. We show that fluorescent FABPs possess a unique, conserved tripeptide Gly-Pro-Pro sequence motif, which is not found in non-fluorescent fatty acid binding proteins. This motif arose from a duplication event of the FABP brain isoforms and was under strong purifying selection, leading to the classification of this new FP family. Residues adjacent to the motif are under strong positive selection, suggesting a further refinement of the eel protein's fluorescent properties. We present a phylogenetic reconstruction of this emerging FP family and describe additional fluorescent FABP members from groups of distantly related eels. The elucidation of this class of fish FPs with diverse properties provides new templates for the development of protein-based fluorescent tools. The evolutionary adaptation from fatty acid-binding proteins to fluorescent fatty acid-binding proteins raises intrigue as to the functional role of bright green fluorescence in this cryptic genus of reclusive eels that inhabit a blue, nearly monochromatic, marine environment.
Conflict of interest statement
Figures


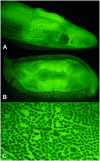
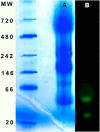

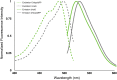


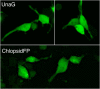
References
-
- Tyler JE, Smith R.C. (1970) Measurements of Spectral Irradiance Underwater New York: Gordon and Breach Science Publishers.
-
- Beebe W, 1935. Half Mile Down. Lane John, The Bodley Head, London. (1935) Half Mile Down. London: John Lane and the Bodley Head.
-
- Shimomura O, Johnson FH, Saiga Y (1962) Extraction, purification and properties of aequorin, a bioluminescent protein from the luminous hydromedusan, Aequorea. J Cell Comp Physiol 59: 223–239. - PubMed
-
- Matz MV, Fradkov AF, Labas YA, Savitsky AP, Zaraisky AG, Markelov ML,. (1999) Fluorescent proteins from nonbioluminescent Anthozoa species. Nat Biotechnol 17: 969–973. - PubMed
-
- Gruber DF, Kao HT, Janoschka S, Tsai J, Pieribone VA (2008) Patterns of fluorescent protein expression in Scleractinian corals. Biol Bull 215: 143–154. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

