Multiple Introduction and Naturally Occuring Drug Resistance of HCV among HIV-Infected Intravenous Drug Users in Yunnan: An Origin of China's HIV/HCV Epidemics
- PMID: 26562015
- PMCID: PMC4642981
- DOI: 10.1371/journal.pone.0142543
Multiple Introduction and Naturally Occuring Drug Resistance of HCV among HIV-Infected Intravenous Drug Users in Yunnan: An Origin of China's HIV/HCV Epidemics
Abstract
Background: The human immunodeficiency virus 1 (HIV-1) epidemic in China historically stemmed from intravenous drug users (IDUs) in Yunnan. Due to a shared transmission route, hepatitis C virus (HCV)/HIV-1 co-infection is common. Here, we investigated HCV genetic characteristics and baseline drug resistance among HIV-infected IDUs in Yunnan.
Methods: Blood samples of 432 HIV-1/HCV co-infected IDUs were collected from January to June 2014 in six prefectures of Yunnan Province. Partial E1E2 and NS5B genes were sequenced. Phylogenetic, evolutionary and genotypic drug resistance analyses were performed.
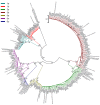
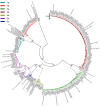
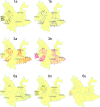
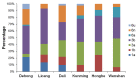
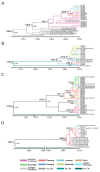
Results: Among the 293 specimens successfully genotyped, seven subtypes were identified, including subtypes 3b (37.9%, 111/293), 3a (21.8%, 64/293), 6n (14.0%, 41/293), 1b (10.6%, 31/293), 1a (8.2%, 24/293), 6a (5.1%, 15/293) and 6u (2.4%, 7/293). The distribution of HCV subtypes was mostly related to geographic location. Subtypes 3b, 3a, and 6n were detected in all six prefectures, however, the other four subtypes were detected only in parts of the six prefectures. Phylogeographic analyses indicated that 6n, 1a and 6u originated in the western prefecture (Dehong) and spread eastward and showed genetic relatedness with those detected in Burmese. However, 6a originated in the southeast prefectures (Honghe and Wenshan) bordering Vietnam and was transmitted westward. These subtypes exhibited different evolutionary rates (between 4.35×10-4 and 2.38×10-3 substitutions site-1 year-1) and times of most recent common ancestor (tMRCA, between 1790.3 and 1994.6), suggesting that HCV was multiply introduced into Yunnan. Naturally occurring resistance-associated mutations (C316N, A421V, C445F, I482L, V494A, and V499A) to NS5B polymerase inhibitors were detected in direct-acting antivirals (DAAs)-naïve IDUs.
Conclusion: This work reveals the temporal-spatial distribution of HCV subtypes and baseline HCV drug resistance among HIV-infected IDUs in Yunnan. The findings enhance our understanding of the characteristics and evolution of HCV in IDUs and are valuable for developing HCV prevention and management strategies for this population.
Conflict of interest statement
Figures
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

