SNPase-ARMS qPCR: Ultrasensitive Mutation-Based Detection of Cell-Free Tumor DNA in Melanoma Patients
- PMID: 26562020
- PMCID: PMC4642939
- DOI: 10.1371/journal.pone.0142273
SNPase-ARMS qPCR: Ultrasensitive Mutation-Based Detection of Cell-Free Tumor DNA in Melanoma Patients
Abstract
Cell-free circulating tumor DNA in the plasma of cancer patients has become a common point of interest as indicator of therapy options and treatment response in clinical cancer research. Especially patient- and tumor-specific single nucleotide variants that accurately distinguish tumor DNA from wild type DNA are promising targets. The reliable detection and quantification of these single-base DNA variants is technically challenging. Currently, a variety of techniques is applied, with no apparent "gold standard". Here we present a novel qPCR protocol that meets the conditions of extreme sensitivity and specificity that are required for detection and quantification of tumor DNA. By consecutive application of two polymerases, one of them designed for extreme base-specificity, the method reaches unprecedented sensitivity and specificity. Three qPCR assays were tested with spike-in experiments, specific for point mutations BRAF V600E, PTEN T167A and NRAS Q61L of melanoma cell lines. It was possible to detect down to one copy of tumor DNA per reaction (Poisson distribution), at a background of up to 200 000 wild type DNAs. To prove its clinical applicability, the method was successfully tested on a small cohort of BRAF V600E positive melanoma patients.
Conflict of interest statement
Figures


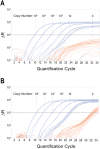
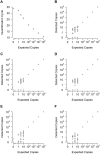

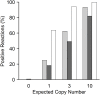
References
-
- Haber DA, Velculescu VE. Blood-based analyses of cancer: circulating tumor cells and circulating tumor DNA. Cancer Discov. 2014;4(6):650–61. 10.1158/2159-8290.CD-13-1014 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

