The Cotton WRKY Gene GhWRKY41 Positively Regulates Salt and Drought Stress Tolerance in Transgenic Nicotiana benthamiana
- PMID: 26562293
- PMCID: PMC4643055
- DOI: 10.1371/journal.pone.0143022
The Cotton WRKY Gene GhWRKY41 Positively Regulates Salt and Drought Stress Tolerance in Transgenic Nicotiana benthamiana
Erratum in
-
Correction: The Cotton WRKY Gene GhWRKY41 Positively Regulates Salt and Drought Stress Tolerance in Transgenic Nicotiana benthamiana.PLoS One. 2016 Jun 2;11(6):e0157026. doi: 10.1371/journal.pone.0157026. eCollection 2016. PLoS One. 2016. PMID: 27253990 Free PMC article.
Abstract
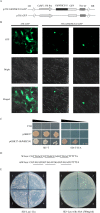
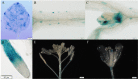
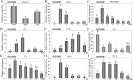
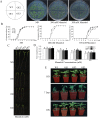
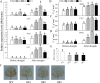
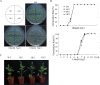
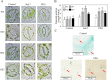
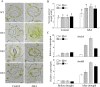
WRKY transcription factors constitute a very large family of proteins in plants and participate in modulating plant biological processes, such as growth, development and stress responses. However, the exact roles of WRKY proteins are unclear, particularly in non-model plants. In this study, Gossypium hirsutum WRKY41 (GhWRKY41) was isolated and transformed into Nicotiana benthamiana. Our results showed that overexpression of GhWRKY41 enhanced the drought and salt stress tolerance of transgenic Nicotiana benthamiana. The transgenic plants exhibited lower malondialdehyde content and higher antioxidant enzyme activity, and the expression of antioxidant genes was upregulated in transgenic plants exposed to osmotic stress. A β-glucuronidase (GUS) staining assay showed that GhWRKY41 was highly expressed in the stomata when plants were exposed to osmotic stress, and plants overexpressing GhWRKY41 exhibited enhanced stomatal closure when they were exposed to osmotic stress. Taken together, our findings demonstrate that GhWRKY41 may enhance plant tolerance to stress by functioning as a positive regulator of stoma closure and by regulating reactive oxygen species (ROS) scavenging and the expression of antioxidant genes.
Conflict of interest statement
Figures


References
-
- Shinozaki K, Yamaguchi-Shinozaki K, Seki M (2003) Regulatory network of gene expression in the drought and cold stress responses. Curr Opin Plant Biol 6:410–417. - PubMed
-
- Jones RJ, Mansfield TA (1970) Suppression of Stomatal Opening in Leaves Treated with Abscisic Acid. J Exp Bot 21:714–719.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

