Diversity of Mycobacterium tuberculosis across Evolutionary Scales
- PMID: 26562841
- PMCID: PMC4642946
- DOI: 10.1371/journal.ppat.1005257
Diversity of Mycobacterium tuberculosis across Evolutionary Scales
Abstract
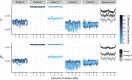
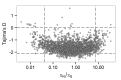
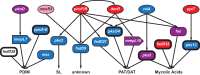
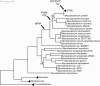
Tuberculosis (TB) is a global public health emergency. Increasingly drug resistant strains of Mycobacterium tuberculosis (M.tb) continue to emerge and spread, highlighting adaptability of this pathogen. Most studies of M.tb evolution have relied on 'between-host' samples, in which each person with TB is represented by a single M.tb isolate. However, individuals with TB commonly harbor populations of M.tb numbering in the billions. Here, we use analyses of M.tb genomic data from within and between hosts to gain insight into influences shaping genetic diversity of this pathogen. We find that the amount of M.tb genetic diversity harbored by individuals with TB can vary dramatically, likely as a function of disease severity. Surprisingly, we did not find an appreciable impact of TB treatment on M.tb diversity. In examining genomic data from M.tb samples within and between hosts with TB, we find that genes involved in the regulation, synthesis, and transportation of immunomodulatory cell envelope lipids appear repeatedly in the extremes of various statistical measures of diversity. Many of these genes have been identified as possible targets of selection in other studies employing different methods and data sets. Taken together, these observations suggest that M.tb cell envelope lipids are targets of selection within hosts. Many of these lipids are specific to pathogenic mycobacteria and, in some cases, human-pathogenic mycobacteria. We speculate that rapid adaptation of cell envelope lipids is facilitated by functional redundancy, flexibility in their metabolism, and their roles mediating interactions with the host.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- WHO | Global tuberculosis report 2014. In: WHO [Internet]. [cited 6 Jan 2015]. http://www.who.int/tb/publications/global_report/en/
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

