Differentiation of human ESCs to retinal ganglion cells using a CRISPR engineered reporter cell line
- PMID: 26563826
- PMCID: PMC4643248
- DOI: 10.1038/srep16595
Differentiation of human ESCs to retinal ganglion cells using a CRISPR engineered reporter cell line
Abstract
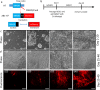
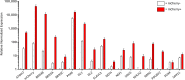
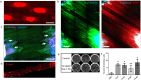
Retinal ganglion cell (RGC) injury and cell death from glaucoma and other forms of optic nerve disease is a major cause of irreversible vision loss and blindness. Human pluripotent stem cell (hPSC)-derived RGCs could provide a source of cells for the development of novel therapeutic molecules as well as for potential cell-based therapies. In addition, such cells could provide insights into human RGC development, gene regulation, and neuronal biology. Here, we report a simple, adherent cell culture protocol for differentiation of hPSCs to RGCs using a CRISPR-engineered RGC fluorescent reporter stem cell line. Fluorescence-activated cell sorting of the differentiated cultures yields a highly purified population of cells that express a range of RGC-enriched markers and exhibit morphological and physiological properties typical of RGCs. Additionally, we demonstrate that aligned nanofiber matrices can be used to guide the axonal outgrowth of hPSC-derived RGCs for in vitro optic nerve-like modeling. Lastly, using this protocol we identified forskolin as a potent promoter of RGC differentiation.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

