Efficient Generation of Myostatin Mutations in Pigs Using the CRISPR/Cas9 System
- PMID: 26564781
- PMCID: PMC4643223
- DOI: 10.1038/srep16623
Efficient Generation of Myostatin Mutations in Pigs Using the CRISPR/Cas9 System
Abstract
Genetically modified pigs are increasingly used for biomedical and agricultural applications. The efficient CRISPR/Cas9 gene editing system holds great promise for the generation of gene-targeting pigs without selection marker genes. In this study, we aimed to disrupt the porcine myostatin (MSTN) gene, which functions as a negative regulator of muscle growth. The transfection efficiency of porcine fetal fibroblasts (PFFs) was improved to facilitate the targeting of Cas9/gRNA. We also demonstrated that Cas9/gRNA can induce non-homologous end-joining (NHEJ), long fragment deletions/inversions and homology-directed repair (HDR) at the MSTN locus of PFFs. Single-cell MSTN knockout colonies were used to generate cloned pigs via somatic cell nuclear transfer (SCNT), which resulted in 8 marker-gene-free cloned pigs with biallelic mutations. Some of the piglets showed obvious intermuscular grooves and enlarged tongues, which are characteristic of the double muscling (DM) phenotype. The protein level of MSTN was decreased in the mutant cloned pigs compared with the wild-type controls, and the mRNA levels of MSTN and related signaling pathway factors were also analyzed. Finally, we carefully assessed off-target mutations in the cloned pigs. The gene editing platform used in this study can efficiently generate genetically modified pigs with biological safety.
Figures
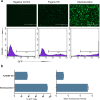
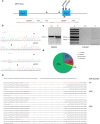

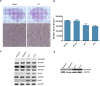
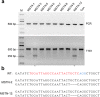
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

