Understanding the Basics of NGS: From Mechanism to Variant Calling
- PMID: 26566462
- PMCID: PMC4633438
- DOI: 10.1007/s40142-015-0076-8
Understanding the Basics of NGS: From Mechanism to Variant Calling
Abstract
Identifying disease-causing mutations in DNA has long been the goal of genetic medicine. In the last decade, the toolkit for discovering DNA variants has undergone rapid evolution: mutations that were historically discovered by analog approaches like Sanger sequencing and multiplex ligation-dependent probe amplification ("MLPA") can now be decoded from a digital signal with next-generation sequencing ("NGS"). Given the explosive growth of NGS-based tests in the clinic, it is of the utmost importance that medical practitioners have a fundamental understanding of the newest NGS methodologies. To that end, here we provide a very basic overview of how NGS works, with particular emphasis on the close resemblance between the underlying chemistry of Sanger sequencing and NGS. Using a pair of simple analogies, we develop an intuitive framework for understanding how high-confidence detection of single-nucleotide polymorphisms, indels, and large deletions/duplications is possible with NGS alone.
Keywords: Del/dup calling; Next-generation sequencing (NGS); Read depth; SNP/indel calling; Variant calling.
Figures
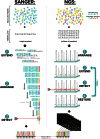


References
-
- National Research Council (U.S.), Committee on Mapping and Sequencing the Human Genome, Alberts B. Report of the Committee on Mapping and Sequencing the Human Genome. Washington, DC: National Academies; 1988.
-
- •• Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. Nature Publishing Group; 2001;409:860–921. This landmark paper announced the release of the nearly complete human genome sequence. It describes the technical aspects of sequencing and assembling the genome, and it presents analysis of the properties and large-scale trends in the sequence. - PubMed
-
- National Human Genome Research Institute. The Human Genome Project Completion: frequently asked questions [Internet]. genome.gov. https://www.genome.gov/11006943. Cited 29 April 2015.
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
