SCRaMbLE generates designed combinatorial stochastic diversity in synthetic chromosomes
- PMID: 26566658
- PMCID: PMC4691749
- DOI: 10.1101/gr.193433.115
SCRaMbLE generates designed combinatorial stochastic diversity in synthetic chromosomes
Abstract
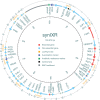
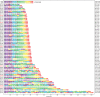
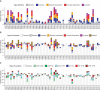
Synthetic chromosome rearrangement and modification by loxP-mediated evolution (SCRaMbLE) generates combinatorial genomic diversity through rearrangements at designed recombinase sites. We applied SCRaMbLE to yeast synthetic chromosome arm synIXR (43 recombinase sites) and then used a computational pipeline to infer or unscramble the sequence of recombinations that created the observed genomes. Deep sequencing of 64 synIXR SCRaMbLE strains revealed 156 deletions, 89 inversions, 94 duplications, and 55 additional complex rearrangements; several duplications are consistent with a double rolling circle mechanism. Every SCRaMbLE strain was unique, validating the capability of SCRaMbLE to explore a diverse space of genomes. Rearrangements occurred exclusively at designed loxPsym sites, with no significant evidence for ectopic rearrangements or mutations involving synthetic regions, the 99% nonsynthetic nuclear genome, or the mitochondrial genome. Deletion frequencies identified genes required for viability or fast growth. Replacement of 3' UTR by non-UTR sequence had surprisingly little effect on fitness. SCRaMbLE generates genome diversity in designated regions, reveals fitness constraints, and should scale to simultaneous evolution of multiple synthetic chromosomes.
© 2016 Shen et al.; Published by Cold Spring Harbor Laboratory Press.
Figures





References
-
- Cello J, Paul AV, Wimmer E. 2002. Chemical synthesis of poliovirus cDNA: generation of infectious virus in the absence of natural template. Science 297: 1016–1018. - PubMed
-
- Dymond JS. 2011. “Construction and characterization of the partially synthetic yeasts Saccharomyces cerevisiae synIXR and semi-synVIL.” Dissertation, The Johns Hopkins University, Baltimore, MD.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
