An active site rearrangement within the Tetrahymena group I ribozyme releases nonproductive interactions and allows formation of catalytic interactions
- PMID: 26567314
- PMCID: PMC4691833
- DOI: 10.1261/rna.053710.115
An active site rearrangement within the Tetrahymena group I ribozyme releases nonproductive interactions and allows formation of catalytic interactions
Abstract
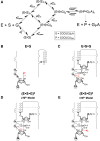
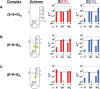
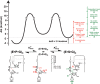
Biological catalysis hinges on the precise structural integrity of an active site that binds and transforms its substrates and meeting this requirement presents a unique challenge for RNA enzymes. Functional RNAs, including ribozymes, fold into their active conformations within rugged energy landscapes that often contain misfolded conformers. Here we uncover and characterize one such "off-pathway" species within an active site after overall folding of the ribozyme is complete. The Tetrahymena group I ribozyme (E) catalyzes cleavage of an oligonucleotide substrate (S) by an exogenous guanosine (G) cofactor. We tested whether specific catalytic interactions with G are present in the preceding E•S•G and E•G ground-state complexes. We monitored interactions with G via the effects of 2'- and 3'-deoxy (-H) and -amino (-NH(2)) substitutions on G binding. These and prior results reveal that G is bound in an inactive configuration within E•G, with the nucleophilic 3'-OH making a nonproductive interaction with an active site metal ion termed MA and with the adjacent 2'-OH making no interaction. Upon S binding, a rearrangement occurs that allows both -OH groups to contact a different active site metal ion, termed M(C), to make what are likely to be their catalytic interactions. The reactive phosphoryl group on S promotes this change, presumably by repositioning the metal ions with respect to G. This conformational transition demonstrates local rearrangements within an otherwise folded RNA, underscoring RNA's difficulty in specifying a unique conformation and highlighting Nature's potential to use local transitions of RNA in complex function.
Keywords: RNA catalysis; active site; conformational change; metal ion; noncoding RNA.
© 2015 Sengupta et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures




Similar articles
-
Probing the role of metal ions in RNA catalysis: kinetic and thermodynamic characterization of a metal ion interaction with the 2'-moiety of the guanosine nucleophile in the Tetrahymena group I ribozyme.Biochemistry. 1999 Aug 24;38(34):10958-75. doi: 10.1021/bi990388e. Biochemistry. 1999. PMID: 10460151
-
A rearrangement of the guanosine-binding site establishes an extended network of functional interactions in the Tetrahymena group I ribozyme active site.Biochemistry. 2010 Mar 30;49(12):2753-62. doi: 10.1021/bi902200n. Biochemistry. 2010. PMID: 20175542 Free PMC article.
-
Protonated 2'-aminoguanosine as a probe of the electrostatic environment of the active site of the Tetrahymena group I ribozyme.Biochemistry. 1999 Aug 24;38(34):10976-88. doi: 10.1021/bi9903897. Biochemistry. 1999. PMID: 10460152
-
Maximizing RNA folding rates: a balancing act.RNA. 2000 Jun;6(6):790-4. doi: 10.1017/s1355838200000522. RNA. 2000. PMID: 10864039 Free PMC article. Review.
-
Folding mechanisms of group I ribozymes: role of stability and contact order.Biochem Soc Trans. 2002 Nov;30(Pt 6):1166-9. doi: 10.1042/bst0301166. Biochem Soc Trans. 2002. PMID: 12440997 Review.
Cited by
-
Enhancement of RNA/Ligand Association Kinetics via an Electrostatic Anchor.Biochemistry. 2019 Jun 18;58(24):2760-2768. doi: 10.1021/acs.biochem.9b00231. Epub 2019 Jun 3. Biochemistry. 2019. PMID: 31117387 Free PMC article.
-
Differential Assembly of Catalytic Interactions within the Conserved Active Sites of Two Ribozymes.PLoS One. 2016 Aug 8;11(8):e0160457. doi: 10.1371/journal.pone.0160457. eCollection 2016. PLoS One. 2016. PMID: 27501145 Free PMC article.
-
Who stole the proton? Suspect general base guanine found with a smoking gun in the pistol ribozyme.Org Biomol Chem. 2022 Aug 10;20(31):6219-6230. doi: 10.1039/d2ob00234e. Org Biomol Chem. 2022. PMID: 35452066 Free PMC article.
-
Hidden intermediates in Mango III RNA aptamer folding revealed by pressure perturbation.Biophys J. 2022 Feb 1;121(3):421-429. doi: 10.1016/j.bpj.2021.12.037. Epub 2021 Dec 28. Biophys J. 2022. PMID: 34971617 Free PMC article.
References
-
- Abramovitz DL, Friedman RA, Pyle AM. 1996. Catalytic role of 2′-hydroxyl groups within a group II intron active site. Science 271: 1410–1413. - PubMed
-
- Adams PL, Stahley MR, Kosek AB, Wang J, Strobel SA. 2004. Crystal structure of a self-splicing group I intron with both exons. Nature 430: 45–50. - PubMed
-
- Bartley LE, Zhuang X, Das R, Chu S, Herschlag D. 2003. Exploration of the transition state for tertiary structure formation between an RNA helix and a large structured RNA. J Mol Biol 328: 1011–1026. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
