Genome-wide Screening of Regulators of Catalase Expression: ROLE OF A TRANSCRIPTION COMPLEX AND HISTONE AND tRNA MODIFICATION COMPLEXES ON ADAPTATION TO STRESS
- PMID: 26567340
- PMCID: PMC4705398
- DOI: 10.1074/jbc.M115.696658
Genome-wide Screening of Regulators of Catalase Expression: ROLE OF A TRANSCRIPTION COMPLEX AND HISTONE AND tRNA MODIFICATION COMPLEXES ON ADAPTATION TO STRESS
Abstract
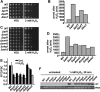
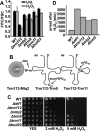
In response to environmental cues, the mitogen-activated protein kinase Sty1-driven signaling cascade activates hundreds of genes to induce a robust anti-stress cellular response in fission yeast. Thus, upon stress imposition Sty1 transiently accumulates in the nucleus where it up-regulates transcription through the Atf1 transcription factor. Several regulators of transcription and translation have been identified as important to mount an integral response to oxidative stress, such as the Spt-Ada-Gcn5-acetyl transferase or Elongator complexes, respectively. With the aim of identifying new regulators of this massive gene expression program, we have used a GFP-based protein reporter and screened a fission yeast deletion collection using flow cytometry. We find that the levels of catalase fused to GFP, both before and after a threat of peroxides, are altered in hundreds of strains lacking components of chromatin modifiers, transcription complexes, and modulators of translation. Thus, the transcription elongation complex Paf1, the histone methylase Set1-COMPASS, and the translation-related Trm112 dimers are all involved in full expression of Ctt1-GFP and in wild-type tolerance to peroxides.
Keywords: Schizosaccharomyces pombe; catalase; gene expression; stress response; transfer RNA (tRNA).
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

