Role of Titin Missense Variants in Dilated Cardiomyopathy
- PMID: 26567375
- PMCID: PMC4845231
- DOI: 10.1161/JAHA.115.002645
Role of Titin Missense Variants in Dilated Cardiomyopathy
Abstract
Background: The titin gene (TTN) encodes the largest human protein, which plays a central role in sarcomere organization and passive myocyte stiffness. TTN truncating mutations cause dilated cardiomyopathy (DCM); however, the role of TTN missense variants in DCM has been difficult to elucidate because of the presence of background TTN variation.
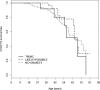
Methods and results: A cohort of 147 DCM index subjects underwent DNA sequencing for 313 TTN exons covering the N2B and N2BA cardiac isoforms of TTN. Of the 348 missense variants, we identified 44 "severe" rare variants by using a bioinformatic filtering process in 37 probands. Of these, 5 probands were double heterozygotes (additional variant in another DCM gene) and 7 were compound heterozygotes (2 TTN "severe" variants). Segregation analysis allowed the classification of the "severe" variants into 5 "likely" (cosegregating), 5 "unlikely" (noncosegregating), and 34 "possibly" (where family structure precluded segregation analysis) disease-causing variants. Patients with DCM carrying "likely" or "possibly" pathogenic TTN "severe" variants did not show a different outcome compared with "unlikely" and noncarriers of a "severe" TTN variant. However, the "likely" and "possibly" disease-causing variants were overrepresented in the C-zone of the A-band region of the sarcomere.
Conclusions: TTN missense variants are common and present a challenge for bioinformatic classification, especially when informative families are not available. Although DCM patients carrying bioinformatically "severe" TTN variants do not appear to have a worse clinical course than noncarriers, the nonrandom distribution of "likely" and "possibly" disease-causing variants suggests a potential biological role for some TTN missense variants.
Keywords: cardiomyopathy; cardiovascular genetics; dilated cardiomyopathy; heart failure; missense variants.
© 2015 The Authors. Published on behalf of the American Heart Association, Inc., by Wiley Blackwell.
Figures




References
-
- Jefferies JL, Towbin JA. Dilated cardiomyopathy. Lancet. 2010;375:752–762. - PubMed
-
- Herman DS, Lam L, Taylor MR, Wang L, Teekakirikul P, Christodoulou D, Conner L, DePalma SR, McDonough B, Sparks E, Teodorescu DL, Cirino AL, Banner NR, Pennell DJ, Graw S, Merlo M, Di Lenarda A, Sinagra G, Bos JM, Ackerman MJ, Mitchell RN, Murry CE, Lakdawala NK, Ho CY, Barton PJ, Cook SA, Mestroni L, Seidman JG, Seidman CE. Truncations of titin causing dilated cardiomyopathy. N Engl J Med. 2012;366:619–628. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HL062881/HL/NHLBI NIH HHS/United States
- R01 HL147064/HL/NHLBI NIH HHS/United States
- R01 HL109209/HL/NHLBI NIH HHS/United States
- N01-HV-48194/HV/NHLBI NIH HHS/United States
- R01 HL69071/HL/NHLBI NIH HHS/United States
- R01HL109209/HL/NHLBI NIH HHS/United States
- R01 HL069071/HL/NHLBI NIH HHS/United States
- M01 RR000051/RR/NCRR NIH HHS/United States
- R01 HL116906/HL/NHLBI NIH HHS/United States
- 3R01HL109209-01A1S1/HL/NHLBI NIH HHS/United States
- R01 HL115988/HL/NHLBI NIH HHS/United States
- UL1 RR025780/RR/NCRR NIH HHS/United States
- HL062881/HL/NHLBI NIH HHS/United States
- 3R01HL109209-01A1S/HL/NHLBI NIH HHS/United States
- UL1 TR001082/TR/NCATS NIH HHS/United States
LinkOut - more resources
Full Text Sources

