Upper Palaeolithic genomes reveal deep roots of modern Eurasians
- PMID: 26567969
- PMCID: PMC4660371
- DOI: 10.1038/ncomms9912
Upper Palaeolithic genomes reveal deep roots of modern Eurasians
Abstract
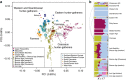
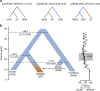
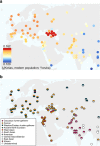
We extend the scope of European palaeogenomics by sequencing the genomes of Late Upper Palaeolithic (13,300 years old, 1.4-fold coverage) and Mesolithic (9,700 years old, 15.4-fold) males from western Georgia in the Caucasus and a Late Upper Palaeolithic (13,700 years old, 9.5-fold) male from Switzerland. While we detect Late Palaeolithic-Mesolithic genomic continuity in both regions, we find that Caucasus hunter-gatherers (CHG) belong to a distinct ancient clade that split from western hunter-gatherers ∼45 kya, shortly after the expansion of anatomically modern humans into Europe and from the ancestors of Neolithic farmers ∼25 kya, around the Last Glacial Maximum. CHG genomes significantly contributed to the Yamnaya steppe herders who migrated into Europe ∼3,000 BC, supporting a formative Caucasus influence on this important Early Bronze age culture. CHG left their imprint on modern populations from the Caucasus and also central and south Asia possibly marking the arrival of Indo-Aryan languages.
Figures


References
-
- Skoglund P. et al. Genomic diversity and admixture differs for stone-age Scandinavian foragers and farmers. Science 344, 747–750 (2014). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

