Intra-tumor genetic heterogeneity in rectal cancer
- PMID: 26568296
- PMCID: PMC4695247
- DOI: 10.1038/labinvest.2015.131
Intra-tumor genetic heterogeneity in rectal cancer
Abstract
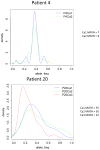
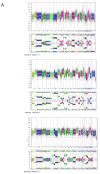
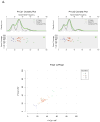
Colorectal cancer arises in part from the cumulative effects of multiple gene lesions. Recent studies in selected cancer types have revealed significant intra-tumor genetic heterogeneity and highlighted its potential role in disease progression and resistance to therapy. We hypothesized the existence of significant intra-tumor genetic heterogeneity in rectal cancers involving variations in localized somatic mutations and copy number abnormalities. Two or three spatially disparate regions from each of six rectal tumors were dissected and subjected to the next-generation whole-exome DNA sequencing, Oncoscan SNP arrays, and targeted confirmatory sequencing and analysis. The resulting data were integrated to define subclones using SciClone. Mutant-allele tumor heterogeneity (MATH) scores, mutant allele frequency correlation, and mutation percent concordance were calculated, and copy number analysis including measurement of correlation between samples was performed. Somatic mutations profiles in individual cancers were similar to prior studies, with some variants found in previously reported significantly mutated genes and many patient-specific mutations in each tumor. Significant intra-tumor heterogeneity was identified in the spatially disparate regions of individual cancers. All tumors had some heterogeneity but the degree of heterogeneity was quite variable in the samples studied. We found that 67-97% of exonic somatic mutations were shared among all regions of an individual's tumor. The SciClone computational method identified 2-8 shared and unshared subclones in the spatially disparate areas in each tumor. MATH scores ranged from 7 to 41. Allele frequency correlation scores ranged from R(2)=0.69-0.96. Measurements of correlation between samples for copy number changes varied from R(2)=0.74-0.93. All tumors had some heterogeneity, but the degree was highly variable in the samples studied. The occurrence of significant intra-tumor heterogeneity may allow selected tumors to have a genetic reservoir to draw from in their evolutionary response to therapy and other challenges.
Figures



References
-
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012 Jan-Feb;62(1):10–29. - PubMed
-
- van Gijn W, Marijnen CA, Nagtegaal ID, Kranenbarg EM, Putter H, Wiggers T, et al. Preoperative radiotherapy combined with total mesorectal excision for resectable rectal cancer: 12-year follow-up of the multicentre, randomised controlled TME trial. Lancet Oncol. 2011 Jun;12(6):575–82. - PubMed
-
- Habr-Gama A, de Souza PM, Ribeiro U, Jr, Nadalin W, Gansl R, Sousa AH, Jr, et al. Low rectal cancer: impact of radiation and chemotherapy on surgical treatment. Dis Colon Rectum. 1998 Sep;41(9):1087–96. - PubMed
-
- Garcia-Aguilar J, Hernandez de Anda E, Sirivongs P, Lee SH, Madoff RD, Rothenberger DA. A pathologic complete response to preoperative chemoradiation is associated with lower local recurrence and improved survival in rectal cancer patients treated by mesorectal excision. Dis Colon Rectum. 2003 Mar;46(3):298–304. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

