Recent advances in the biosynthesis of unusual polyketide synthase substrates
- PMID: 26571143
- PMCID: PMC4742410
- DOI: 10.1039/c5np00112a
Recent advances in the biosynthesis of unusual polyketide synthase substrates
Abstract
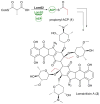
This highlight provides an overview of recent advances in understanding the diversity of polyketide synthase (PKS) substrate building blocks. Substrates functioning as starter units and extender units contribute significantly to the chemical complexity and structural diversity exhibited by this class of natural products. This article complements and extends upon the current comprehensive reviews that have been published on these two topics (Moore and Hertweck, Nat. Prod. Rep., 2002, 19, 70; Chan et al., Nat. Prod. Rep., 2009, 1, 90; Wilson and Moore, Nat. Prod. Rep., 2012, 29, 72).
Figures
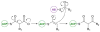

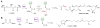




References
-
- Staunton J, Weissman KJ. Nat Prod Rep. 2001;18:380–416. - PubMed
-
- Fischbach MA, Walsh CT. Chem Rev. 2006;106:3468–3496. - PubMed
-
- Hertweck C, Luzhetskyy A, Rebets Y, Bechthold A. Nat Prod Rep. 2007;24:162–190. - PubMed
-
- Austin MB, Noel JP. Nat Prod Rep. 2003;20:79–110. - PubMed
-
- Hertweck C. Angew Chem, Int Ed. 2009;48:4688–4716. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

