Mutations in NONO lead to syndromic intellectual disability and inhibitory synaptic defects
- PMID: 26571461
- PMCID: PMC5392243
- DOI: 10.1038/nn.4169
Mutations in NONO lead to syndromic intellectual disability and inhibitory synaptic defects
Abstract
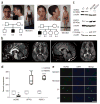
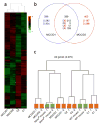
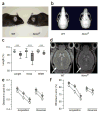
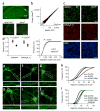
The NONO protein has been characterized as an important transcriptional regulator in diverse cellular contexts. Here we show that loss of NONO function is a likely cause of human intellectual disability and that NONO-deficient mice have cognitive and affective deficits. Correspondingly, we find specific defects at inhibitory synapses, where NONO regulates synaptic transcription and gephyrin scaffold structure. Our data identify NONO as a possible neurodevelopmental disease gene and highlight the key role of the DBHS protein family in functional organization of GABAergic synapses.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

