SS18-SSX, the Oncogenic Fusion Protein in Synovial Sarcoma, Is a Cellular Context-Dependent Epigenetic Modifier
- PMID: 26571495
- PMCID: PMC4646489
- DOI: 10.1371/journal.pone.0142991
SS18-SSX, the Oncogenic Fusion Protein in Synovial Sarcoma, Is a Cellular Context-Dependent Epigenetic Modifier
Abstract
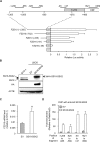
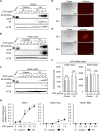
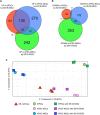
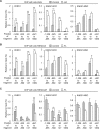
The prevalence and specificity of unique fusion oncogenes are high in a number of soft tissue sarcomas (STSs). The close relationship between fusion genes and clinicopathological features suggests that a correlation may exist between the function of fusion proteins and cellular context of the cell-of-origin of each tumor. However, most STSs are origin-unknown tumors and this issue has not yet been investigated in detail. In the present study, we examined the effects of the cellular context on the function of the synovial sarcoma (SS)-specific fusion protein, SS18-SSX, using human pluripotent stem cells (hPSCs) containing the drug-inducible SS18-SSX gene. We selected the neural crest cell (NCC) lineage for the first trial of this system, induced SS18-SSX at various differentiation stages from PSCs to NCC-derived mesenchymal stromal cells (MSCs), and compared its biological effects on each cell type. We found that the expression of FZD10, identified as an SS-specific gene, was induced by SS18-SSX at the PSC and NCC stages, but not at the MSC stage. This stage-specific induction of FZD10 correlated with stage-specific changes in histone marks associated with the FZD10 locus and also with the loss of the BAF47 protein, a member of the SWI/SNF chromatin-remodeling complex. Furthermore, the global gene expression profile of hPSC-derived NCCs was the closest to that of SS cell lines after the induction of SS18-SSX. These results clearly demonstrated that the cellular context is an important factor in the function of SS18-SSX as an epigenetic modifier.
Conflict of interest statement
Figures

References
-
- Eifert C, Powers RS (2012) From cancer genomes to oncogenic drivers, tumour dependencies and therapeutic targets. Nat Rev Cancer 12: 572–578. - PubMed
-
- Zucman J, Delattre O, Desmaze C, Epstein AL, Stenman G, Speleman F, et al. (1993) EWS and ATF-1 gene fusion induced by t(12;22) translocation in malignant melanoma of soft parts. Nat Genet 4: 341–345. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

