The Deubiquitinating Enzyme UBPY Is Required for Lysosomal Biogenesis and Productive Autophagy in Drosophila
- PMID: 26571504
- PMCID: PMC4646453
- DOI: 10.1371/journal.pone.0143078
The Deubiquitinating Enzyme UBPY Is Required for Lysosomal Biogenesis and Productive Autophagy in Drosophila
Abstract
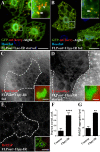
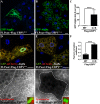
Autophagy is a catabolic process that delivers cytoplasmic components to the lysosomes. Protein modification by ubiquitination is involved in this pathway: it regulates the stability of autophagy regulators such as BECLIN-1 and it also functions as a tag targeting specific substrates to autophagosomes. In order to identify deubiquitinating enzymes (DUBs) involved in autophagy, we have performed a genetic screen in the Drosophila larval fat body. This screen identified Uch-L3, Usp45, Usp12 and Ubpy. In this paper, we show that Ubpy loss of function results in the accumulation of autophagosomes due to a blockade of the autophagy flux. Furthermore, analysis by electron and confocal microscopy of Ubpy-depleted fat body cells revealed altered lysosomal morphology, indicating that Ubpy inactivation affects lysosomal maintenance and/or biogenesis. Lastly, we have shown that shRNA mediated inactivation of UBPY in HeLa cells affects autophagy in a different way: in UBPY-depleted HeLa cells autophagy is deregulated.
Conflict of interest statement
Figures




Similar articles
-
A deubiquitinating enzyme UBPY regulates the level of protein ubiquitination on endosomes.Traffic. 2006 Aug;7(8):1017-31. doi: 10.1111/j.1600-0854.2006.00452.x. Epub 2006 Jun 12. Traffic. 2006. PMID: 16771824
-
A functional endosomal pathway is necessary for lysosome biogenesis in Drosophila.BMC Cell Biol. 2016 Nov 16;17(1):36. doi: 10.1186/s12860-016-0115-7. BMC Cell Biol. 2016. PMID: 27852225 Free PMC article.
-
The deubiquitinating enzyme USP36 controls selective autophagy activation by ubiquitinated proteins.Autophagy. 2012 May 1;8(5):767-79. doi: 10.4161/auto.19381. Epub 2012 May 1. Autophagy. 2012. PMID: 22622177
-
Ubiquitin-specific protease 8 (USP8/UBPy): a prototypic multidomain deubiquitinating enzyme with pleiotropic functions.Biochem Soc Trans. 2019 Dec 20;47(6):1867-1879. doi: 10.1042/BST20190527. Biochem Soc Trans. 2019. PMID: 31845722 Free PMC article. Review.
-
Regulation of endocytic sorting by ESCRT-DUB-mediated deubiquitination.Cell Biochem Biophys. 2011 Jun;60(1-2):39-46. doi: 10.1007/s12013-011-9181-9. Cell Biochem Biophys. 2011. PMID: 21448666 Review.
Cited by
-
The deubiquitinase Leon/USP5 interacts with Atg1/ULK1 and antagonizes autophagy.Cell Death Dis. 2023 Aug 22;14(8):540. doi: 10.1038/s41419-023-06062-x. Cell Death Dis. 2023. PMID: 37607937 Free PMC article.
-
Degradation of arouser by endosomal microautophagy is essential for adaptation to starvation in Drosophila.Life Sci Alliance. 2020 Dec 14;4(2):e202000965. doi: 10.26508/lsa.202000965. Print 2021 Feb. Life Sci Alliance. 2020. PMID: 33318080 Free PMC article.
-
Deubiquitylase USP12 induces pro-survival autophagy and bortezomib resistance in multiple myeloma by stabilizing HMGB1.Oncogene. 2022 Feb;41(9):1298-1308. doi: 10.1038/s41388-021-02167-9. Epub 2022 Jan 7. Oncogene. 2022. PMID: 34997217
-
The deubiquitinase USP45 inhibits autophagy through actin regulation by Coronin 1B.J Cell Biol. 2025 May 5;224(5):e202407014. doi: 10.1083/jcb.202407014. Epub 2025 Mar 11. J Cell Biol. 2025. PMID: 40067150 Free PMC article.
-
Investigation of Chemical Composition and Biological Activities of Ajuga pyramidalis-Isolation of Iridoids and Phenylethanoid Glycosides.Metabolites. 2023 Jan 14;13(1):128. doi: 10.3390/metabo13010128. Metabolites. 2023. PMID: 36677053 Free PMC article.
References
-
- Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402(6762):672–6. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

