Complementing the inflammasome
- PMID: 26572245
- PMCID: PMC4717242
- DOI: 10.1111/imm.12556
Complementing the inflammasome
Abstract
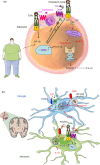
The innate immune system is an ancient surveillance system able to sense microbial invaders as well as aberrations in normal cell function. No longer viewed as a static and non-specific part of immunity, the innate immune system employs a plethora of specialized pattern recognition sensors to monitor and achieve homeostasis; these include the Toll-like receptors, the retinoic acid-inducible gene-like receptors, the nucleotide-binding oligomerization domain receptors (NLRs), the C-type lectins and the complement system. In order to increase specificity and diversity, innate immunity uses homotypic and heterotypic associations among these different components. Multi-molecular assemblies are formed both on the cell surface and in the cytosol to respond to pathogen and danger signals. Diverse, but tailored, responses to a changing environment are orchestrated depending on the the nature of the challenge and the repertoire of interacting receptors and components available in the sensing cell. It is now emerging that innate immunity operates a system of 'checks and balances' where interaction among the sensors is key in maintaining normal cell function. Complement sits at the heart of this alarm system and it is becoming apparent that it is capable of interacting with all the other pathways to effect a tailored immune response. In this review, we will focus on complement interactions with NLRs, the so-called 'inflammasomes', describing the molecular mechanisms that have been revealed so far and discussing the circumstantial evidence that exists for these interactions in disease states.
Keywords: cell surface molecules; complement; inflammasome; inflammation; innate lymphoid cells.
© 2015 John Wiley & Sons Ltd.
Figures

References
-
- Gaboriaud C, Thielens N, Gregory L, Rossi V, Fontecilla‐Camps J, Arlaud G. Structure and activation of the C1 complex of complement: unraveling the puzzle. Trends Immunol 2004; 25:368–73. - PubMed
-
- Chen C, Wallis R. Two mechanisms for mannose‐binding protein modulation of the activity of its associated serine proteases. J Biol Chem 2004; 279:26058–65. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

