Identification of the meiotic toolkit in diatoms and exploration of meiosis-specific SPO11 and RAD51 homologs in the sexual species Pseudo-nitzschia multistriata and Seminavis robusta
- PMID: 26572248
- PMCID: PMC4647503
- DOI: 10.1186/s12864-015-1983-5
Identification of the meiotic toolkit in diatoms and exploration of meiosis-specific SPO11 and RAD51 homologs in the sexual species Pseudo-nitzschia multistriata and Seminavis robusta
Erratum in
-
Correction to: Identification of the meiotic toolkit in diatoms and exploration of meiosis-specific SPO11 and RAD51 homologs in the sexual species Pseudo-nitzschia multistriata and Seminavis robusta.BMC Genomics. 2019 Jul 5;20(1):544. doi: 10.1186/s12864-019-5942-4. BMC Genomics. 2019. PMID: 31277569 Free PMC article.
Abstract
Background: Sexual reproduction is an obligate phase in the life cycle of most eukaryotes. Meiosis varies among organisms, which is reflected by the variability of the gene set associated to the process. Diatoms are unicellular organisms that belong to the stramenopile clade and have unique life cycles that can include a sexual phase.
Results: The exploration of five diatom genomes and one diatom transcriptome led to the identification of 42 genes potentially involved in meiosis. While these include the majority of known meiosis-related genes, several meiosis-specific genes, including DMC1, could not be identified. Furthermore, phylogenetic analyses supported gene identification and revealed ancestral loss and recent expansion in the RAD51 family in diatoms. The two sexual species Pseudo-nitzschia multistriata and Seminavis robusta were used to explore the expression of meiosis-related genes: RAD21, SPO11-2, RAD51-A, RAD51-B and RAD51-C were upregulated during meiosis, whereas other paralogs in these families showed no differential expression patterns, suggesting that they may play a role during vegetative divisions. An almost identical toolkit is shared among Pseudo-nitzschia multiseries and Fragilariopsis cylindrus, as well as two species for which sex has not been observed, Phaeodactylum tricornutum and Thalassiosira pseudonana, suggesting that these two may retain a facultative sexual phase.
Conclusions: Our results reveal the conserved meiotic toolkit in six diatom species and indicate that Stramenopiles share major modifications of canonical meiosis processes ancestral to eukaryotes, with important divergences in each Kingdom.
Figures
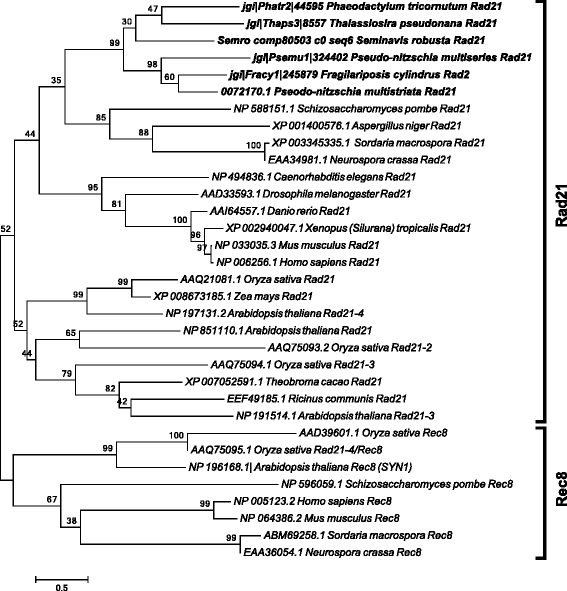
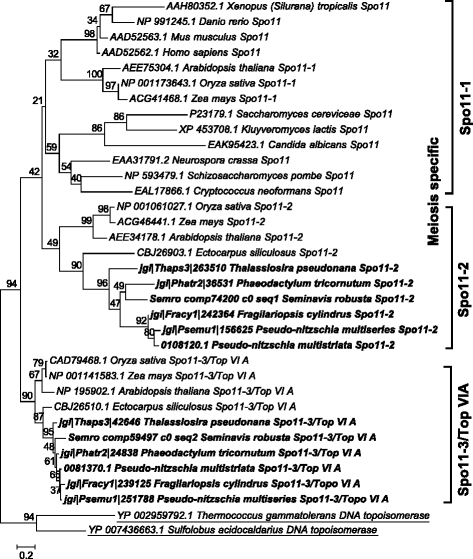
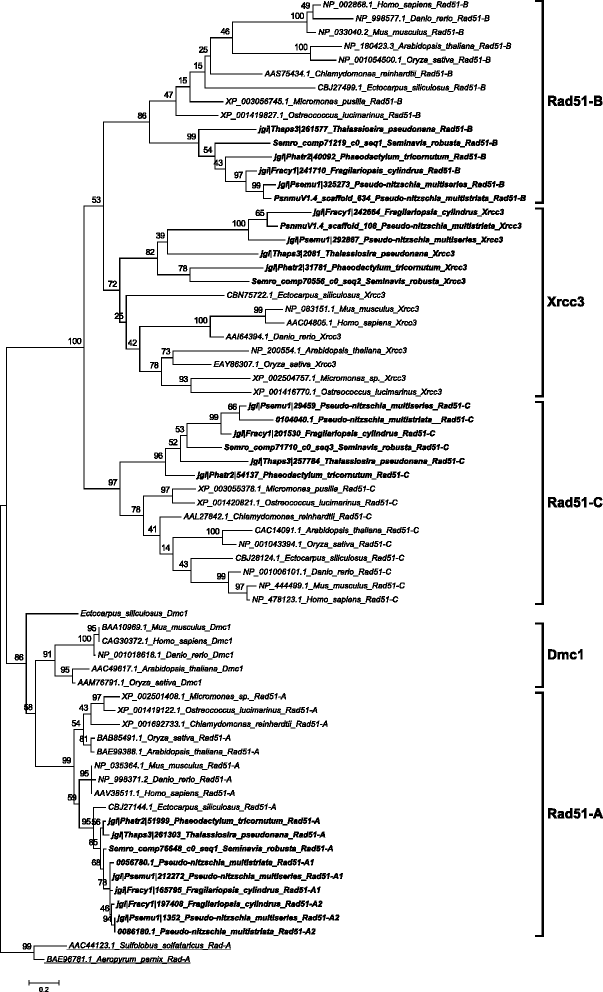

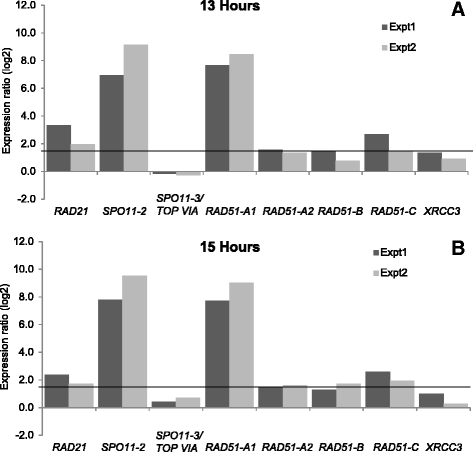
References
-
- Cavalier-Smith T. Origins of the machinery of recombination and sex. Heredity (Edinb) 2002;88:125–41. - PubMed
-
- Zimmer C. On the origin of sexual reproduction. Science. 2009;324:1254–6. - PubMed
-
- Schurko AM, Logsdon JM. Using a meiosis detection toolkit to investigate ancient asexual “scandals” and the evolution of sex. BioEssays. 2008;30:579–89. - PubMed
-
- Ramesh MA, Malik SB, Logsdon JM. A phylogenomic inventory of meiotic genes: Evidence for sex in Giardia and an early eukaryotic origin of meiosis. Curr Biol. 2005;15:185–91. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

