Sample storage conditions significantly influence faecal microbiome profiles
- PMID: 26572876
- PMCID: PMC4648095
- DOI: 10.1038/srep16350
Sample storage conditions significantly influence faecal microbiome profiles
Abstract
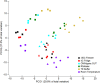
Sequencing-based studies of the human faecal microbiota are increasingly common. Appropriate storage of sample material is essential to avoid the introduction of post-collection bias in microbial community composition. Rapid freezing to -80 °C is commonly considered to be best-practice. However, this is not feasible in many studies, particularly those involving sample collection in participants' homes. We determined the extent to which a range of stabilisation and storage strategies maintained the composition of faecal microbial community structure relative to freezing to -80 °C. Refrigeration at 4 °C, storage at ambient temperature, and the use of several common preservative buffers (RNAlater, OMNIgene.GUT, Tris-EDTA) were assessed relative to freezing. Following 72 hours of storage, faecal microbial composition was assessed by 16 S rRNA amplicon sequencing. Refrigeration was associated with no significant alteration in faecal microbiota diversity or composition. However, samples stored using other conditions showed substantial divergence compared to -80 °C control samples. Aside from refrigeration, the use of OMNIgene.GUT resulted in the least alteration, while the greatest change was seen in samples stored in Tris-EDTA buffer. The commercially available OMNIgene.GUT kit may provide an important alternative where refrigeration and cold chain transportation is not available.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

