Mango (Mangifera indica L.) germplasm diversity based on single nucleotide polymorphisms derived from the transcriptome
- PMID: 26573148
- PMCID: PMC4647706
- DOI: 10.1186/s12870-015-0663-6
Mango (Mangifera indica L.) germplasm diversity based on single nucleotide polymorphisms derived from the transcriptome
Abstract
Background: Germplasm collections are an important source for plant breeding, especially in fruit trees which have a long duration of juvenile period. Thus, efforts have been made to study the diversity of fruit tree collections. Even though mango is an economically important crop, most of the studies on diversity in mango collections have been conducted with a small number of genetic markers.
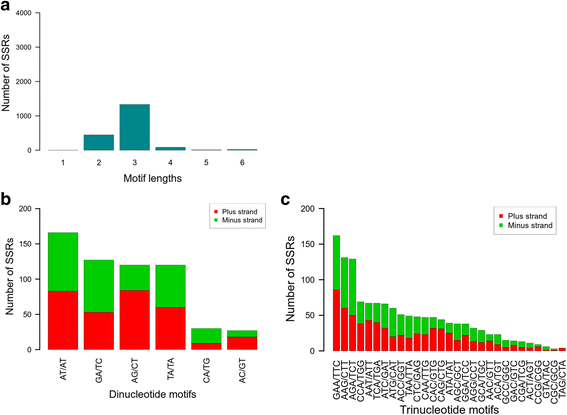
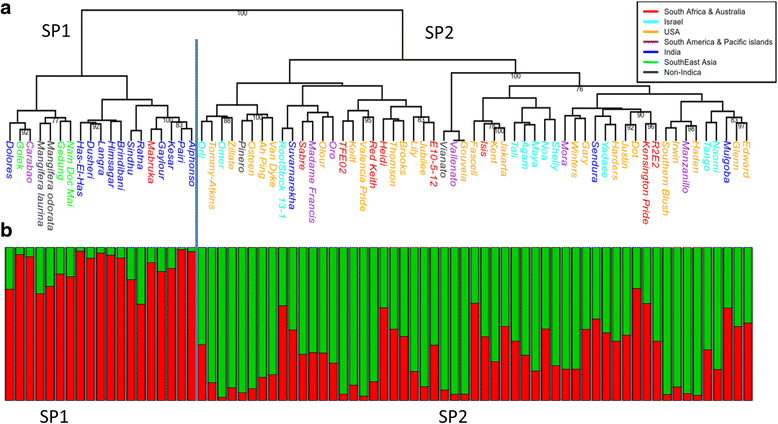
Results: We describe a de novo transcriptome assembly from mango cultivar 'Keitt'. Variation discovery was performed using Illumina resequencing of 'Keitt' and 'Tommy Atkins' cultivars identified 332,016 single-nucleotide polymorphisms (SNPs) and 1903 simple-sequence repeats (SSRs). Most of the SSRs (70.1%) were of trinucleotide with the preponderance of motif (GGA/AAG)n and only 23.5% were di-nucleotide SSRs with the mostly of (AT/AT)n motif. Further investigation of the diversity in the Israeli mango collection was performed based on a subset of 293 SNPs. Those markers have divided the Israeli mango collection into two major groups: one group included mostly mango accessions from Southeast Asia (Malaysia, Thailand, Indonesia) and India and the other with mainly of Floridian and Israeli mango cultivars. The latter group was more polymorphic (FS=-0.1 on the average) and was more of an admixture than the former group. A slight population differentiation was detected (FST=0.03), suggesting that if the mango accessions of the western world apparently was originated from Southeast Asia, as has been previously suggested, the duration of cultivation was not long enough to develop a distinct genetic background.
Conclusions: Whole-transcriptome reconstruction was used to significantly broaden the mango's genetic variation resources, i.e., SNPs and SSRs. The set of SNP markers described in this study is novel. A subset of SNPs was sampled to explore the Israeli mango collection and most of them were polymorphic in many mango accessions. Therefore, we believe that these SNPs will be valuable as they recapitulate and strengthen the history of mango diversity.
Figures
Similar articles
-
The 'Tommy Atkins' mango genome reveals candidate genes for fruit quality.BMC Plant Biol. 2021 Feb 22;21(1):108. doi: 10.1186/s12870-021-02858-1. BMC Plant Biol. 2021. PMID: 33618672 Free PMC article.
-
Genetic Diversity and Fingerprinting of 231 Mango Germplasm Using Genome SSR Markers.Int J Mol Sci. 2024 Dec 19;25(24):13625. doi: 10.3390/ijms252413625. Int J Mol Sci. 2024. PMID: 39769387 Free PMC article.
-
Population structure and genetic diversity of mango (Mangifera indica L.) germplasm resources as revealed by single-nucleotide polymorphism markers.Front Plant Sci. 2024 Jul 3;15:1328126. doi: 10.3389/fpls.2024.1328126. eCollection 2024. Front Plant Sci. 2024. PMID: 39022611 Free PMC article.
-
Advances in Physiological, Transcriptomic, Proteomic, Metabolomic, and Molecular Genetic Approaches for Enhancing Mango Fruit Quality.J Agric Food Chem. 2023 Jan 11;71(1):20-34. doi: 10.1021/acs.jafc.2c05958. Epub 2022 Dec 27. J Agric Food Chem. 2023. PMID: 36573879 Review.
-
Multifaceted Health Benefits of Mangifera indica L. (Mango): The Inestimable Value of Orchards Recently Planted in Sicilian Rural Areas.Nutrients. 2017 May 20;9(5):525. doi: 10.3390/nu9050525. Nutrients. 2017. PMID: 28531110 Free PMC article. Review.
Cited by
-
Complete mitochondrial genomes of three Mangifera species, their genomic structure and gene transfer from chloroplast genomes.BMC Genomics. 2022 Feb 19;23(1):147. doi: 10.1186/s12864-022-08383-1. BMC Genomics. 2022. PMID: 35183120 Free PMC article.
-
In vitro antifungal activity of dimethyl trisulfide against Colletotrichum gloeosporioides from mango.World J Microbiol Biotechnol. 2019 Dec 12;36(1):4. doi: 10.1007/s11274-019-2781-z. World J Microbiol Biotechnol. 2019. PMID: 31832786
-
Genetic diversity of avocado (Persea americana Mill.) germplasm using pooled sequencing.BMC Genomics. 2019 May 15;20(1):379. doi: 10.1186/s12864-019-5672-7. BMC Genomics. 2019. PMID: 31092188 Free PMC article.
-
Comparative Assessment of SSR and RAPD markers for genetic diversity in some Mango cultivars.PeerJ. 2023 Sep 28;11:e15722. doi: 10.7717/peerj.15722. eCollection 2023. PeerJ. 2023. PMID: 37790610 Free PMC article.
-
Transcriptome Dynamics in Mango Fruit Peel Reveals Mechanisms of Chilling Stress.Front Plant Sci. 2016 Oct 20;7:1579. doi: 10.3389/fpls.2016.01579. eCollection 2016. Front Plant Sci. 2016. PMID: 27812364 Free PMC article.
References
-
- Bompard J. Taxonomy and systematics. In: Litz RE, editor. The mango: Botany, production and uses. Wallingford: CAB International; 2009. pp. 19–41.
-
- Mukherjee S, Litz R. Introduction: botany and importance. In: Litz RE, editor. The mango: Botany, production and uses. 2. Wallingford: CAB international; 2009. pp. 1–18.
-
- Olano CT, Schnell RJ, Quintanilla WE, Campbell RJ. Pedigree analysis of Florida mango cultivars. Proc Fla State Hort Soc. 2005;118:192–197.
-
- Bally IE, Lu P, Johnson P. Mango breeding. In: Jain SM, Priyadarshan PM, editors. Breeding plantation tree crops: tropical species. New York: Springer; 2009. pp. 51–82.
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

