Gene expression and functional annotation of human choroid plexus epithelium failure in Alzheimer's disease
- PMID: 26573292
- PMCID: PMC4647590
- DOI: 10.1186/s12864-015-2159-z
Gene expression and functional annotation of human choroid plexus epithelium failure in Alzheimer's disease
Abstract
Background: Alzheimer's disease (AD) is the most common form of dementia. AD has a multifactorial disease etiology and is currently untreatable. Multiple genes and molecular mechanisms have been implicated in AD, including ß-amyloid deposition in the brain, neurofibrillary tangle accumulation of hyper-phosphorylated Tau, synaptic failure, oxidative stress and inflammation. Relatively little is known about the role of the blood-brain barriers, especially the blood-cerebrospinal fluid barrier (BCSFB), in AD. The BCSFB is involved in cerebrospinal fluid (CSF) production, maintenance of brain homeostasis and neurodegenerative disorders.
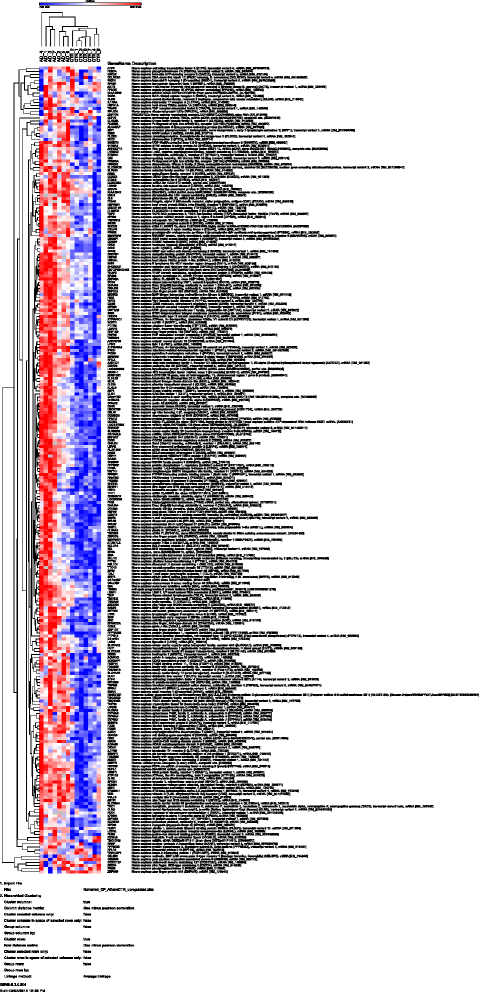
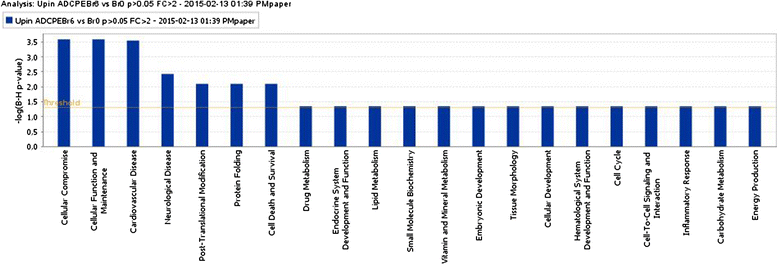
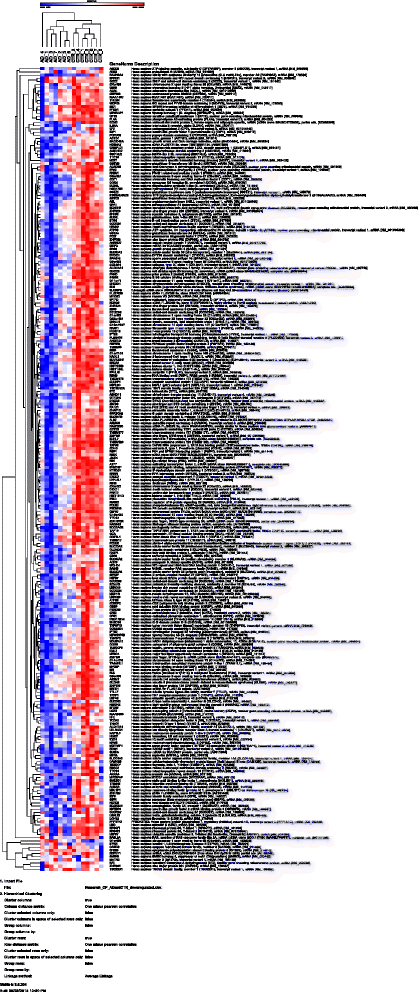
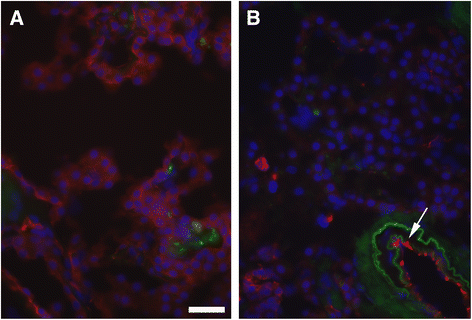
Results: Using an Agilent platform with common reference design, we performed a large scale gene expression analysis and functional annotation of the Choroid Plexus Epithelium (CPE), which forms the BCSFB. We obtained 2 groups of freshly frozen Choroid Plexus (CP) of 7 human donor brains each, with and without AD: Braak stages (0-1) and (5-6). We cut CP cryo-sections and isolated RNA from cresyl-violet stained, laser dissected CPE cells. Gene expression results were analysed with T-tests (R) and the knowledge-database Ingenuity. We found statistically significantly altered gene expression data sets, biological functions, canonical pathways, molecular networks and functionalities in AD-affected CPE. We observed specific cellular changes due to increased oxidative stress, such as the unfolded protein response, E1F2 and NRF2 signalling and the protein ubiquitin pathway. Most likely, the AD-affected BCSFB barrier becomes more permeable due to downregulation of CLDN5. Finally, our data also predicted down regulation of the glutathione mediated detoxification pathway and the urea cycle in the AD CPE, which suggest that the CPE sink action may be impaired. Remarkably, the expression of a number of genes known to be involved in AD, such as APP, PSEN1, PSEN2, TTR and CLU is moderate to high and remains stable in both healthy and affected CPE. Literature labelling of our new functional molecular networks confirmed multiple previous (molecular) observations in the AD literature and revealed many new ones.
Conclusions: We conclude that CPE failure in AD exists. Combining our data with those of the literature, we propose the following chronological and overlapping chain of events: increased Aß burden on CPE; increased oxidative stress in CPE; despite continuous high expression of TTR: decreased capability of CPE to process amyloid; (pro-) inflammatory and growth factor signalling by CPE; intracellular ubiquitin involvement, remodelling of CPE tight junctions and, finally, cellular atrophy. Our data corroborates the hypothesis that increased BCSFB permeability, especially loss of selective CLDN5-mediated paracellular transport, altered CSF production and CPE sink action, as well as loss of CPE mediated macrophage recruitment contribute to the pathogenesis of AD.
Figures

References
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

