The recruitment of chromatin modifiers by long noncoding RNAs: lessons from PRC2
- PMID: 26574518
- PMCID: PMC4647455
- DOI: 10.1261/rna.053918.115
The recruitment of chromatin modifiers by long noncoding RNAs: lessons from PRC2
Abstract
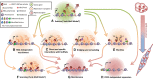
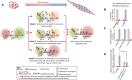
Polycomb repressive complex-2 (PRC2) is a histone methyltransferase required for epigenetic silencing during development and cancer. Among chromatin modifying factors shown to be recruited and regulated by long noncoding RNAs (lncRNAs), PRC2 is one of the most studied. Mammalian PRC2 binds thousands of RNAs in vivo, and it is becoming a model system for the recruitment of chromatin modifying factors by RNA. Yet, well-defined PRC2-binding motifs within target RNAs have been elusive. From the protein side, PRC2 RNA-binding subunits contain no known RNA-binding domains, complicating functional studies. Here we provide a critical review of existing models for the recruitment of PRC2 to chromatin by RNAs. This discussion may also serve researchers who are studying the recruitment of other chromatin modifiers by lncRNAs.
Keywords: PRC2; RNA–protein interaction; epigenetic silencing; histone modification; long noncoding RNAs.
© 2015 Davidovich and Cech; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures

References
-
- Agback P, Baumann H, Knapp S, Ladenstein R, Härd T. 1998. Architecture of nonspecific protein-DNA interactions in the Sso7d-DNA complex. Nat Struct Biol 5: 579–584. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
