Genetic Variation for Thermotolerance in Lettuce Seed Germination Is Associated with Temperature-Sensitive Regulation of ETHYLENE RESPONSE FACTOR1 (ERF1)
- PMID: 26574598
- PMCID: PMC4704578
- DOI: 10.1104/pp.15.01251
Genetic Variation for Thermotolerance in Lettuce Seed Germination Is Associated with Temperature-Sensitive Regulation of ETHYLENE RESPONSE FACTOR1 (ERF1)
Abstract
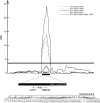
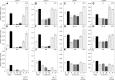
Seeds of most lettuce (Lactuca sativa) cultivars are susceptible to thermoinhibition, or failure to germinate at temperatures above approximately 28°C, creating problems for crop establishment in the field. Identifying genes controlling thermoinhibition would enable the development of cultivars lacking this trait and, therefore, being less sensitive to high temperatures during planting. Seeds of a primitive accession (PI251246) of lettuce exhibited high-temperature germination capacity up to 33°C. Screening a recombinant inbred line population developed from PI215246 and cv Salinas identified a major quantitative trait locus (Htg9.1) from PI251246 associated with the high-temperature germination phenotype. Further genetic analyses discovered a tight linkage of the Htg9.1 phenotype with a specific DNA marker (NM4182) located on a single genomic sequence scaffold. Expression analyses of the 44 genes encoded in this genomic region revealed that only a homolog of Arabidopsis (Arabidopsis thaliana) ETHYLENE RESPONSE FACTOR1 (termed LsERF1) was differentially expressed between PI251246 and cv Salinas seeds imbibed at high temperature (30°C). LsERF1 belongs to a large family of transcription factors associated with the ethylene-signaling pathway. Physiological assays of ethylene synthesis, response, and action in parental and near-isogenic Htg9.1 genotypes strongly implicate LsERF1 as the gene responsible for the Htg9.1 phenotype, consistent with the established role for ethylene in germination thermotolerance of Compositae seeds. Expression analyses of genes associated with the abscisic acid and gibberellin biosynthetic pathways and results of biosynthetic inhibitor and hormone response experiments also support the hypothesis that differential regulation of LsERF1 expression in PI251246 seeds elevates their upper temperature limit for germination through interactions among pathways regulated by these hormones. Our results support a model in which LsERF1 acts through the promotion of gibberellin biosynthesis to counter the inhibitory effects of abscisic acid and, therefore, promote germination at high temperatures.
© 2016 American Society of Plant Biologists. All Rights Reserved.
Figures





References
-
- Argyris J, Truco MJ, Ochoa O, Knapp SJ, Still DW, Lenssen GM, Schut JW, Michelmore RW, Bradford KJ (2005) Quantitative trait loci associated with seed and seedling traits in Lactuca. Theor Appl Genet 111: 1365–1376 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

