Proteorhodopsin light-enhanced growth linked to vitamin-B1 acquisition in marine Flavobacteria
- PMID: 26574687
- PMCID: PMC5029205
- DOI: 10.1038/ismej.2015.196
Proteorhodopsin light-enhanced growth linked to vitamin-B1 acquisition in marine Flavobacteria
Abstract
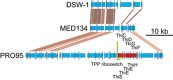
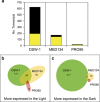
Proteorhodopsins (PR) are light-driven proton pumps widely distributed in bacterioplankton. Although they have been thoroughly studied for more than a decade, it is still unclear how the proton motive force (pmf) generated by PR is used in most organisms. Notably, very few PR-containing bacteria show growth enhancement in the light. It has been suggested that the presence of specific functions within a genome may define the different PR-driven light responses. Thus, comparing closely related organisms that respond differently to light is an ideal setup to identify the mechanisms involved in PR light-enhanced growth. Here, we analyzed the transcriptomes of three PR-harboring Flavobacteria strains of the genus Dokdonia: Dokdonia donghaensis DSW-1(T), Dokdonia MED134 and Dokdonia PRO95, grown in identical seawater medium in light and darkness. Although only DSW-1(T) and MED134 showed light-enhanced growth, all strains expressed their PR genes at least 10 times more in the light compared with dark. According to their genomes, DSW-1(T) and MED134 are vitamin-B1 auxotrophs, and their vitamin-B1 TonB-dependent transporters (TBDT), accounted for 10-18% of all pmf-dependent transcripts. In contrast, the expression of vitamin-B1 TBDT was 10 times lower in the prototroph PRO95, whereas its vitamin-B1 synthesis genes were among the highest expressed. Our data suggest that light-enhanced growth in DSW-1(T) and MED134 derives from the use of PR-generated pmf to power the uptake of vitamin-B1, essential for central carbon metabolism, including the TCA cycle. Other pmf-generating mechanisms available in darkness are probably insufficient to power transport of enough vitamin-B1 to support maximum growth of these organisms.
Figures




References
-
- Béjà O, Aravind L, Koonin EV, Suzuki MT, Hadd A, Nguyen LP et al. (2000). Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science 289: 1902–1906. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

