Off-target Effects in CRISPR/Cas9-mediated Genome Engineering
- PMID: 26575098
- PMCID: PMC4877446
- DOI: 10.1038/mtna.2015.37
Off-target Effects in CRISPR/Cas9-mediated Genome Engineering
Abstract
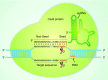
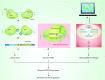
CRISPR/Cas9 is a versatile genome-editing technology that is widely used for studying the functionality of genetic elements, creating genetically modified organisms as well as preclinical research of genetic disorders. However, the high frequency of off-target activity (≥50%)-RGEN (RNA-guided endonuclease)-induced mutations at sites other than the intended on-target site-is one major concern, especially for therapeutic and clinical applications. Here, we review the basic mechanisms underlying off-target cutting in the CRISPR/Cas9 system, methods for detecting off-target mutations, and strategies for minimizing off-target cleavage. The improvement off-target specificity in the CRISPR/Cas9 system will provide solid genotype-phenotype correlations, and thus enable faithful interpretation of genome-editing data, which will certainly facilitate the basic and clinical application of this technology.
Figures
References
-
- Doudna, JA and Charpentier, E (2014). Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346: 1258096. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

