454 Pyrosequencing of Olive (Olea europaea L.) Transcriptome in Response to Salinity
- PMID: 26576008
- PMCID: PMC4648586
- DOI: 10.1371/journal.pone.0143000
454 Pyrosequencing of Olive (Olea europaea L.) Transcriptome in Response to Salinity
Abstract
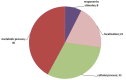
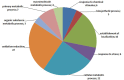
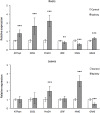
Olive (Olea europaea L.) is one of the most important crops in the Mediterranean region. The expansion of cultivation in areas irrigated with low quality and saline water has negative effects on growth and productivity however the investigation of the molecular basis of salt tolerance in olive trees has been only recently initiated. To this end, we investigated the molecular response of cultivar Kalamon to salinity stress using next-generation sequencing technology to explore the transcriptome profile of olive leaves and roots and identify differentially expressed genes that are related to salt tolerance response. Out of 291,958 obtained trimmed reads, 28,270 unique transcripts were identified of which 35% are annotated, a percentage that is comparable to similar reports on non-model plants. Among the 1,624 clusters in roots that comprise more than one read, 24 were differentially expressed comprising 9 down- and 15 up-regulated genes. Respectively, inleaves, among the 2,642 clusters, 70 were identified as differentially expressed, with 14 down- and 56 up-regulated genes. Using next-generation sequencing technology we were able to identify salt-response-related transcripts. Furthermore we provide an annotated transcriptome of olive as well as expression data, which are both significant tools for further molecular studies in olive.
Conflict of interest statement
Figures
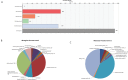
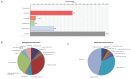
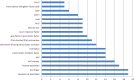
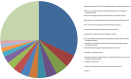



References
-
- Tattini M, Bertoni P, Caselli S. Genotypic responses of olive plants to sodium chloride. J Plant Nutr. 1992;15:1467–1485.
-
- Klein I, Ben-Tal Y, Lavee S, De Malach Y, David I. Saline irrigation of cv. Manzanillo and Uovo di Piccione trees. Acta Hort (ISHS). 1994;356:176–180.
-
- Cresti M, Ciampolini F, Tattini M, Cimato A. Effect of salinity on productivity and oil quality of olive (Olea europaea L.) plants. Advances in Horticultural Science. 1994;8:211–214.
-
- Loreto F, Centritto M, Chartzoulakis K. Photosynthetic limitations in olive cultivars with different sensitivity to salt stress. Plant Cell Environ. 2003;26:595–601.
-
- Tabatabaei SJ. Effects of salinity and N on the growth, photosynthesis and N status of olive (Olea europaea L.) trees. Sci Hortic-Amsterdam. 2006;108:432–438.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

