Structural insights into HetR-PatS interaction involved in cyanobacterial pattern formation
- PMID: 26576507
- PMCID: PMC4649674
- DOI: 10.1038/srep16470
Structural insights into HetR-PatS interaction involved in cyanobacterial pattern formation
Abstract
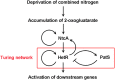
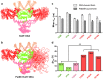
The one-dimensional pattern of heterocyst in the model cyanobacterium Anabaena sp. PCC 7120 is coordinated by the transcription factor HetR and PatS peptide. Here we report the complex structures of HetR binding to DNA, and its hood domain (HetRHood) binding to a PatS-derived hexapeptide (PatS6) at 2.80 and 2.10 Å, respectively. The intertwined HetR dimer possesses a couple of novel HTH motifs, each of which consists of two canonical α-helices in the DNA-binding domain and an auxiliary α-helix from the flap domain of the neighboring subunit. Two PatS6 peptides bind to the lateral clefts of HetRHood, and trigger significant conformational changes of the flap domain, resulting in dissociation of the auxiliary α-helix and eventually release of HetR from the DNA major grove. These findings provide the structural insights into a prokaryotic example of Turing model.
Figures



Similar articles
-
Evidence for direct binding between HetR from Anabaena sp. PCC 7120 and PatS-5.Biochemistry. 2011 Nov 1;50(43):9212-24. doi: 10.1021/bi201226e. Epub 2011 Oct 7. Biochemistry. 2011. PMID: 21942265
-
Differential binding between PatS C-terminal peptide fragments and HetR from Anabaena sp. PCC 7120.Biochemistry. 2012 Mar 27;51(12):2436-42. doi: 10.1021/bi300228n. Epub 2012 Mar 15. Biochemistry. 2012. PMID: 22397695
-
hetR and patS, two genes necessary for heterocyst pattern formation, are widespread in filamentous nonheterocyst-forming cyanobacteria.Microbiology (Reading). 2009 May;155(Pt 5):1418-1426. doi: 10.1099/mic.0.027540-0. Epub 2009 Apr 21. Microbiology (Reading). 2009. PMID: 19383713
-
Heterocyst differentiation and pattern formation in cyanobacteria: a chorus of signals.Mol Microbiol. 2006 Jan;59(2):367-75. doi: 10.1111/j.1365-2958.2005.04979.x. Mol Microbiol. 2006. PMID: 16390435 Review.
-
Heterocyst formation in Anabaena.Curr Opin Microbiol. 1998 Dec;1(6):623-9. doi: 10.1016/s1369-5274(98)80106-9. Curr Opin Microbiol. 1998. PMID: 10066546 Review.
Cited by
-
patD, a Gene Regulated by NtcA, Is Involved in the Optimization of Heterocyst Frequency in the Cyanobacterium Anabaena sp. Strain PCC 7120.J Bacteriol. 2019 Oct 4;201(21):e00457-19. doi: 10.1128/JB.00457-19. Print 2019 Nov 1. J Bacteriol. 2019. PMID: 31405917 Free PMC article.
-
PatU3 plays a central role in coordinating cell division and differentiation in pattern formation of filamentous cyanobacterium Nostoc sp. PCC 7120.Sci China Life Sci. 2023 Dec;66(12):2896-2909. doi: 10.1007/s11427-023-2380-1. Epub 2023 Jul 25. Sci China Life Sci. 2023. PMID: 37505430
-
Role of PatS and cell type on the heterocyst spacing pattern in a filamentous branching cyanobacterium.FEMS Microbiol Lett. 2017 Aug 15;364(15):fnx154. doi: 10.1093/femsle/fnx154. FEMS Microbiol Lett. 2017. PMID: 28859320 Free PMC article.
-
Manipulation of Pattern of Cell Differentiation in a hetR Mutant of Anabaena sp. PCC 7120 by Overexpressing hetZ Alone or with hetP.Life (Basel). 2018 Nov 30;8(4):60. doi: 10.3390/life8040060. Life (Basel). 2018. PMID: 30513635 Free PMC article.
-
Expression from DIF1-motif promoters of hetR and patS is dependent on HetZ and modulated by PatU3 during heterocyst differentiation.PLoS One. 2020 Jul 23;15(7):e0232383. doi: 10.1371/journal.pone.0232383. eCollection 2020. PLoS One. 2020. PMID: 32701963 Free PMC article.
References
-
- Turing A. M. The chemical basis of morphogenesis. Philos. Trans. R. Soc. Lond. 237, 37–72 (1952).
-
- Kondo S. & Miura T. Reaction-diffusion model as a framework for understanding biological pattern formation. Science 329, 1616–1620 (2010). - PubMed
-
- Meinhardt H. Models of biological pattern formation: From elementary steps to the organization of embryonic axes. Curr. Top. Dev. Biol. 81, 1–63 (2008). - PubMed
-
- Haselkorn R. Heterocysts. Annu. Rev. Plant Physio. 29, 319–344 (1978).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

