The tomato mutant ars1 (altered response to salt stress 1) identifies an R1-type MYB transcription factor involved in stomatal closure under salt acclimation
- PMID: 26578112
- PMCID: PMC11388943
- DOI: 10.1111/pbi.12498
The tomato mutant ars1 (altered response to salt stress 1) identifies an R1-type MYB transcription factor involved in stomatal closure under salt acclimation
Abstract
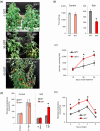
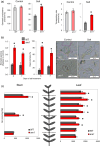
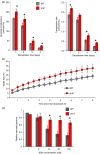
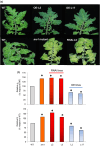
A screening under salt stress conditions of a T-DNA mutant collection of tomato (Solanum lycopersicum L.) led to the identification of the altered response to salt stress 1 (ars1) mutant, which showed a salt-sensitive phenotype. Genetic analysis of the ars1 mutation revealed that a single T-DNA insertion in the ARS1 gene was responsible of the mutant phenotype. ARS1 coded for an R1-MYB type transcription factor and its expression was induced by salinity in leaves. The mutant reduced fruit yield under salt acclimation while in the absence of stress the disruption of ARS1 did not affect this agronomic trait. The stomatal behaviour of ars1 mutant leaves induced higher Na(+) accumulation via the transpiration stream, as the decreases of stomatal conductance and transpiration rate induced by salt stress were markedly lower in the mutant plants. Moreover, the mutation affected stomatal closure in a response mediated by abscisic acid (ABA). The characterization of tomato transgenic lines silencing and overexpressing ARS1 corroborates the role of the gene in regulating the water loss via transpiration under salinity. Together, our results show that ARS1 tomato gene contributes to reduce transpirational water loss under salt stress. Finally, this gene could be interesting for tomato molecular breeding, because its manipulation could lead to improved stress tolerance without yield penalty under optimal culture conditions.
Keywords: MYB transcription factor; Solanum lycopersicum; insertional mutagenesis; salt stress; stomatal aperture; transpiration.
© 2015 Society for Experimental Biology, Association of Applied Biologists and John Wiley & Sons Ltd.
Figures


References
-
- AbuQamar, S. , Luo, H. , Laluk, K. , Mickelbart, M.V. and Mengiste, T. (2009) Crosstalk between biotic and abiotic stress responses in tomato is mediated by the AIM1 transcription factor. Plant J. 58, 347–360. - PubMed
-
- Asins, M.J. , Villalta, I. , Aly, M.M. , Olías, R. , Álvarez De Morales, P.A.Z. , Huertas, R. , Li, J.U.N. et al. (2013) Two closely linked tomato HKT coding genes are positional candidates for the major tomato QTL involved in Na+/K+ homeostasis. Plant Cell Environ. 36, 1171–1191. - PubMed
-
- Atarés, A. , Moyano, E. , Morales, B. , Schleicher, P. , García‐Abellán, J. , Antón, T. , García‐Sogo, B. et al. (2011) An insertional mutagenesis programme with an enhancer trap for the identification and tagging of genes involved in abiotic stress tolerance in the tomato wild‐related species Solanum pennellii . Plant Cell Rep. 30, 1865–1879. - PMC - PubMed
-
- Baulcombe, D.C. , Saunders, G.R. , Bevan, M.W. , Mayo, M.A. and Harrison, B.D. (1986) Expression of biologically active viral satellite RNA from the nuclear genome of transformed plants. Nature, 321, 446–449.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

