Integrating mass spectrometry and genomics for cyanobacterial metabolite discovery
- PMID: 26578313
- PMCID: PMC5065021
- DOI: 10.1007/s10295-015-1705-7
Integrating mass spectrometry and genomics for cyanobacterial metabolite discovery
Abstract
Filamentous marine cyanobacteria produce bioactive natural products with both potential therapeutic value and capacity to be harmful to human health. Genome sequencing has revealed that cyanobacteria have the capacity to produce many more secondary metabolites than have been characterized. The biosynthetic pathways that encode cyanobacterial natural products are mostly uncharacterized, and lack of cyanobacterial genetic tools has largely prevented their heterologous expression. Hence, a combination of cutting edge and traditional techniques has been required to elucidate their secondary metabolite biosynthetic pathways. Here, we review the discovery and refined biochemical understanding of the olefin synthase and fatty acid ACP reductase/aldehyde deformylating oxygenase pathways to hydrocarbons, and the curacin A, jamaicamide A, lyngbyabellin, columbamide, and a trans-acyltransferase macrolactone pathway encoding phormidolide. We integrate into this discussion the use of genomics, mass spectrometric networking, biochemical characterization, and isolation and structure elucidation techniques.
Keywords: Biosynthesis; Cyanobacteria; Genomics; Mass spectrometry; Natural products.
Figures
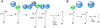




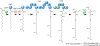
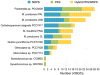


References
-
- Breton RC, Reynolds WF. Using NMR to identify and characterize natural products. Nat Prod Rep. 2013;30:501–524. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

