Eukaryotic Initiation Factor eIFiso4G1 and eIFiso4G2 Are Isoforms Exhibiting Distinct Functional Differences in Supporting Translation in Arabidopsis
- PMID: 26578519
- PMCID: PMC4714232
- DOI: 10.1074/jbc.M115.692939
Eukaryotic Initiation Factor eIFiso4G1 and eIFiso4G2 Are Isoforms Exhibiting Distinct Functional Differences in Supporting Translation in Arabidopsis
Abstract
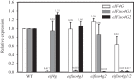
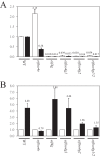
The eukaryotic translation initiation factor (eIF) 4G is required during protein synthesis to promote the assembly of several factors involved in the recruitment of a 40S ribosomal subunit to an mRNA. Although many eukaryotes express two eIF4G isoforms that are highly similar, the eIF4G isoforms in plants, referred to as eIF4G and eIFiso4G, are highly divergent in size, sequence, and domain organization but both can interact with eIF4A, eIF4B, eIF4E isoforms, and the poly(A)-binding protein. Nevertheless, eIF4G and eIFiso4G from wheat exhibit preferences in the mRNAs they translate optimally. For example, mRNA containing the 5'-leader (called Ω) of tobacco mosaic virus preferentially uses eIF4G in wheat germ lysate. In this study, the eIF4G isoform specificity of Ω was used to examine functional differences of the eIF4G isoforms in Arabidopsis. As in wheat, Ω-mediated translation was reduced in an eif4g null mutant. Loss of the eIFiso4G1 isoform, which is similar in sequence to wheat eIFiso4G, did not substantially affect Ω-mediated translation. However, loss of the eIFiso4G2 isoform substantially reduced Ω-mediated translation. eIFiso4G2 is substantially divergent from eIFiso4G1 and is present only in the Brassicaceae, suggesting a recent evolution. eIFiso4G2 isoforms exhibit sequence-specific differences in regions representing partner protein and RNA binding sites. Loss of any eIF4G isoform also resulted in a substantial reduction in reporter transcript level. These results suggest that eIFiso4G2 appeared late in plant evolution and exhibits more functional similarity with eIF4G than with eIFiso4G1 during Ω-mediated translation.
Keywords: RNA; eIFiso4G; eukaryotic translation initiation; eukaryotic translation initiation factor 4G (eIF4G); mRNA stability; protein synthesis; translation; translation initiation factors.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures







Similar articles
-
Plant growth and fertility requires functional interactions between specific PABP and eIF4G gene family members.PLoS One. 2018 Jan 30;13(1):e0191474. doi: 10.1371/journal.pone.0191474. eCollection 2018. PLoS One. 2018. PMID: 29381712 Free PMC article.
-
Competitive and noncompetitive binding of eIF4B, eIF4A, and the poly(A) binding protein to wheat translation initiation factor eIFiso4G.Biochemistry. 2010 Sep 28;49(38):8251-65. doi: 10.1021/bi1008529. Biochemistry. 2010. PMID: 20795652
-
eIF4G, eIFiso4G, and eIF4B bind the poly(A)-binding protein through overlapping sites within the RNA recognition motif domains.J Biol Chem. 2007 Aug 31;282(35):25247-58. doi: 10.1074/jbc.M702193200. Epub 2007 Jul 2. J Biol Chem. 2007. PMID: 17606619
-
Conducting the initiation of protein synthesis: the role of eIF4G.Biol Cell. 2003 May-Jun;95(3-4):141-56. doi: 10.1016/s0248-4900(03)00031-5. Biol Cell. 2003. PMID: 12867079 Review.
-
Unorthodox Mechanisms to Initiate Translation Open Novel Paths for Gene Expression.J Mol Biol. 2020 Dec 4;432(24):166702. doi: 10.1016/j.jmb.2020.10.035. Epub 2020 Nov 7. J Mol Biol. 2020. PMID: 33166539 Review.
Cited by
-
How to Cope with the Challenges of Environmental Stresses in the Era of Global Climate Change: An Update on ROS Stave off in Plants.Int J Mol Sci. 2022 Feb 11;23(4):1995. doi: 10.3390/ijms23041995. Int J Mol Sci. 2022. PMID: 35216108 Free PMC article. Review.
-
Translational gene regulation in plants: A green new deal.Wiley Interdiscip Rev RNA. 2020 Nov;11(6):e1597. doi: 10.1002/wrna.1597. Epub 2020 May 4. Wiley Interdiscip Rev RNA. 2020. PMID: 32367681 Free PMC article. Review.
-
Plant growth and fertility requires functional interactions between specific PABP and eIF4G gene family members.PLoS One. 2018 Jan 30;13(1):e0191474. doi: 10.1371/journal.pone.0191474. eCollection 2018. PLoS One. 2018. PMID: 29381712 Free PMC article.
-
Cryo-EM structure of wheat ribosome reveals unique features of the plant ribosomes.Structure. 2024 May 2;32(5):562-574.e3. doi: 10.1016/j.str.2024.02.006. Epub 2024 Mar 7. Structure. 2024. PMID: 38458197 Free PMC article.
-
Class II members of the poly(A) binding protein family exhibit distinct functions during Arabidopsis growth and development.Translation (Austin). 2017 Feb 17;5(1):e1295129. doi: 10.1080/21690731.2017.1295129. eCollection 2017. Translation (Austin). 2017. PMID: 28702277 Free PMC article.
References
-
- Preiss T., and W. Hentze M. (2003) Starting the protein synthesis machine: eukaryotic translation initiation. Bioessays 25, 1201–1211 - PubMed
-
- Kapp L. D., and Lorsch J. R. (2004) The molecular mechanics of eukaryotic translation. Annu. Rev. Biochem. 73, 657–704 - PubMed
-
- Pestova T. V., Lorsch J. R., and Hellen C.U.T. (2007) The mechanism of translation initiation in eukaryotes. in Translational Control in Biology and Medicine (Mathews M. B., Sonenberg N., and Hershey J. W. B., eds) pp. 87–128, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
-
- Gallie D. R. (2002) Protein-protein interactions required during translation. Plant Mol. Biol. 50, 949–970 - PubMed
-
- Wei C.-C., Balasta M. L., Ren J., and Goss D. J. (1998) Wheat germ poly(A) binding protein enhances the binding affinity of eukaryotic initiation factor 4F and (iso)4F for cap analogues. Biochemistry 37, 1910–1916 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

