Mutations in Replicative Stress Response Pathways Are Associated with S Phase-specific Defects in Nucleotide Excision Repair
- PMID: 26578521
- PMCID: PMC4705374
- DOI: 10.1074/jbc.M115.685883
Mutations in Replicative Stress Response Pathways Are Associated with S Phase-specific Defects in Nucleotide Excision Repair
Abstract
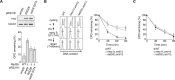
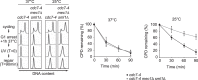
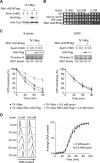
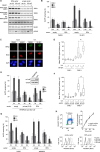
Nucleotide excision repair (NER) is a highly conserved pathway that removes helix-distorting DNA lesions induced by a plethora of mutagens, including UV light. Our laboratory previously demonstrated that human cells deficient in either ATM and Rad3-related (ATR) kinase or translesion DNA polymerase η (i.e. key proteins that promote the completion of DNA replication in response to UV-induced replicative stress) are characterized by profound inhibition of NER exclusively during S phase. Toward elucidating the mechanistic basis of this phenomenon, we developed a novel assay to quantify NER kinetics as a function of cell cycle in the model organism Saccharomyces cerevisiae. Using this assay, we demonstrate that in yeast, deficiency of the ATR homologue Mec1 or of any among several other proteins involved in the cellular response to replicative stress significantly abrogates NER uniquely during S phase. Moreover, initiation of DNA replication is required for manifestation of this defect, and S phase NER proficiency is correlated with the capacity of individual mutants to respond to replicative stress. Importantly, we demonstrate that partial depletion of Rfa1 recapitulates defective S phase-specific NER in wild type yeast; moreover, ectopic RPA1-3 overexpression rescues such deficiency in either ATR- or polymerase η-deficient human cells. Our results strongly suggest that reduction of NER capacity during periods of enhanced replicative stress, ostensibly caused by inordinate sequestration of RPA at stalled DNA replication forks, represents a conserved feature of the multifaceted eukaryotic DNA damage response.
Keywords: DNA replication; Saccharomyces cerevisiae; cell cycle; flow cytometry; nucleotide excision repair; ultraviolet light.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures



References
-
- Schreier W. J., Gilch P., and Zinth W. (2015) Early Events of DNA Photodamage. Annu. Rev. Phys. Chem. 66, 497–519 - PubMed
-
- Marteijn J. A., Lans H., Vermeulen W., and Hoeijmakers J. H. J. (2014) Understanding nucleotide excision repair and its roles in cancer and ageing. Nat. Rev. Mol. Cell Biol. 15, 465–481 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

