Genome-wide binding studies reveal DNA binding specificity mechanisms and functional interplay amongst Forkhead transcription factors
- PMID: 26578569
- PMCID: PMC4770209
- DOI: 10.1093/nar/gkv1120
Genome-wide binding studies reveal DNA binding specificity mechanisms and functional interplay amongst Forkhead transcription factors
Abstract
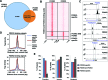
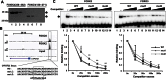
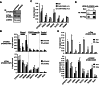
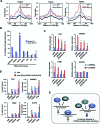
Transcription factors belonging to the same transcription factor families contain very similar DNA binding domains and hence have the potential to bind to related DNA sequences. However, subtle differences in binding specificities can be detected in vitro with the potential to direct specific responses in vivo. Here, we have examined the binding properties of three Forkhead (FOX) transcription factors, FOXK2, FOXO3 and FOXJ3 in vivo. Extensive overlap in chromatin binding is observed, although underlying differential DNA binding specificity can dictate the recruitment of FOXK2 and FOXJ3 to chromatin. However, functionally, FOXO3-dependent gene regulation is generally mediated not through uniquely bound regions but through regions occupied by both FOXK2 and FOXO3 where both factors play a regulatory role. Our data point to a model whereby FOX transcription factors control gene expression through dynamically binding and generating partial occupancy of the same site rather than mutually exclusive binding derived by stable binding of individual FOX proteins.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Jolma A., Yan J., Whitington T., Toivonen J., Nitta K.R., Rastas P., Morgunova E., Enge M., Taipale M., Wei G., et al. DNA-binding specificities of human transcription factors. Cell. 2013;152:327–339. - PubMed
-
- Boros J., Donaldson I.J., O'Donnell A., Odrowaz Z.A., Zeef L., Lupien M., Meyer C.A., Liu X.S., Brown M., Sharrocks A.D. Elucidation of the ELK1 target gene network reveals a role in the coordinate regulation of core components of the gene regulation machinery. Genome Res. 2009;19:1963–1973. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

