Assembly: a resource for assembled genomes at NCBI
- PMID: 26578580
- PMCID: PMC4702866
- DOI: 10.1093/nar/gkv1226
Assembly: a resource for assembled genomes at NCBI
Abstract
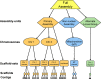
The NCBI Assembly database (www.ncbi.nlm.nih.gov/assembly/) provides stable accessioning and data tracking for genome assembly data. The model underlying the database can accommodate a range of assembly structures, including sets of unordered contig or scaffold sequences, bacterial genomes consisting of a single complete chromosome, or complex structures such as a human genome with modeled allelic variation. The database provides an assembly accession and version to unambiguously identify the set of sequences that make up a particular version of an assembly, and tracks changes to updated genome assemblies. The Assembly database reports metadata such as assembly names, simple statistical reports of the assembly (number of contigs and scaffolds, contiguity metrics such as contig N50, total sequence length and total gap length) as well as the assembly update history. The Assembly database also tracks the relationship between an assembly submitted to the International Nucleotide Sequence Database Consortium (INSDC) and the assembly represented in the NCBI RefSeq project. Users can find assemblies of interest by querying the Assembly Resource directly or by browsing available assemblies for a particular organism. Links in the Assembly Resource allow users to easily download sequence and annotations for current versions of genome assemblies from the NCBI genomes FTP site.
Published by Oxford University Press on behalf of Nucleic Acids Research 2015. This work is written by (a) US Government employee(s) and is in the public domain in the US.
Figures

References
-
- van Dijk E.L., Auger H., Jaszczyszyn Y., Thermes C. Ten years of next-generation sequencing technology. Trends Genet. 2014;30:418–426. - PubMed
-
- O'Leary N., Wright M., Brister J., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D., et al. Reference Sequence (RefSeq) Database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016 doi:10.1093/nar/gkv1189. - PMC - PubMed
-
- Federhen S., Clark K., Barrett T., Parkinson H., Ostell J., Kodama Y., Mashima J., Nakamura Y., Cochrane G., Karsch-Mizrachi I. Toward richer metadata for microbial sequences: replacing strain-level NCBI taxonomy taxids with BioProject, BioSample and Assembly records. Stand. Genomic Sci. 2014;9:1275–1277. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

