Geomicrobiology of sublacustrine thermal vents in Yellowstone Lake: geochemical controls on microbial community structure and function
- PMID: 26579074
- PMCID: PMC4620420
- DOI: 10.3389/fmicb.2015.01044
Geomicrobiology of sublacustrine thermal vents in Yellowstone Lake: geochemical controls on microbial community structure and function
Abstract
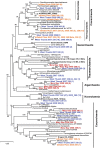
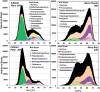
Yellowstone Lake (Yellowstone National Park, WY, USA) is a large high-altitude (2200 m), fresh-water lake, which straddles an extensive caldera and is the center of significant geothermal activity. The primary goal of this interdisciplinary study was to evaluate the microbial populations inhabiting thermal vent communities in Yellowstone Lake using 16S rRNA gene and random metagenome sequencing, and to determine how geochemical attributes of vent waters influence the distribution of specific microorganisms and their metabolic potential. Thermal vent waters and associated microbial biomass were sampled during two field seasons (2007-2008) using a remotely operated vehicle (ROV). Sublacustrine thermal vent waters (circa 50-90°C) contained elevated concentrations of numerous constituents associated with geothermal activity including dissolved hydrogen, sulfide, methane and carbon dioxide. Microorganisms associated with sulfur-rich filamentous "streamer" communities of Inflated Plain and West Thumb (pH range 5-6) were dominated by bacteria from the Aquificales, but also contained thermophilic archaea from the Crenarchaeota and Euryarchaeota. Novel groups of methanogens and members of the Korarchaeota were observed in vents from West Thumb and Elliot's Crater (pH 5-6). Conversely, metagenome sequence from Mary Bay vent sediments did not yield large assemblies, and contained diverse thermophilic and nonthermophilic bacterial relatives. Analysis of functional genes associated with the major vent populations indicated a direct linkage to high concentrations of carbon dioxide, reduced sulfur (sulfide and/or elemental S), hydrogen and methane in the deep thermal ecosystems. Our observations show that sublacustrine thermal vents in Yellowstone Lake support novel thermophilic communities, which contain microorganisms with functional attributes not found to date in terrestrial geothermal systems of YNP.
Keywords: Aquificales; Archaea; hydrogen; metagenome; methane; methanotrophs; sulfide; thermophiles.
Figures



Similar articles
-
Sulfur cycling and host-virus interactions in Aquificales-dominated biofilms from Yellowstone's hottest ecosystems.ISME J. 2022 Mar;16(3):842-855. doi: 10.1038/s41396-021-01132-4. Epub 2021 Oct 14. ISME J. 2022. PMID: 34650231 Free PMC article.
-
Geochemistry and Mixing Drive the Spatial Distribution of Free-Living Archaea and Bacteria in Yellowstone Lake.Front Microbiol. 2016 Feb 29;7:210. doi: 10.3389/fmicb.2016.00210. eCollection 2016. Front Microbiol. 2016. PMID: 26973602 Free PMC article.
-
Microbial communities and chemosynthesis in yellowstone lake sublacustrine hydrothermal vent waters.Front Microbiol. 2011 Jun 13;2:130. doi: 10.3389/fmicb.2011.00130. eCollection 2011. Front Microbiol. 2011. PMID: 21716640 Free PMC article.
-
Metagenomes from high-temperature chemotrophic systems reveal geochemical controls on microbial community structure and function.PLoS One. 2010 Mar 19;5(3):e9773. doi: 10.1371/journal.pone.0009773. PLoS One. 2010. PMID: 20333304 Free PMC article.
-
Metagenome sequence analysis of filamentous microbial communities obtained from geochemically distinct geothermal channels reveals specialization of three aquificales lineages.Front Microbiol. 2013 May 29;4:84. doi: 10.3389/fmicb.2013.00084. eCollection 2013. Front Microbiol. 2013. PMID: 23755042 Free PMC article.
Cited by
-
Functional genes and thermophilic microorganisms responsible for arsenite oxidation from the shallow sediment of an untraversed hot spring outlet.Ecotoxicology. 2017 May;26(4):490-501. doi: 10.1007/s10646-017-1779-2. Epub 2017 Mar 1. Ecotoxicology. 2017. PMID: 28251437
-
Comparing microbial populations from diverse hydrothermal features in Yellowstone National Park: hot springs and mud volcanoes.Front Microbiol. 2024 Jun 27;15:1409664. doi: 10.3389/fmicb.2024.1409664. eCollection 2024. Front Microbiol. 2024. PMID: 38993494 Free PMC article.
-
Sulfur cycling and host-virus interactions in Aquificales-dominated biofilms from Yellowstone's hottest ecosystems.ISME J. 2022 Mar;16(3):842-855. doi: 10.1038/s41396-021-01132-4. Epub 2021 Oct 14. ISME J. 2022. PMID: 34650231 Free PMC article.
-
Geochemistry and Mixing Drive the Spatial Distribution of Free-Living Archaea and Bacteria in Yellowstone Lake.Front Microbiol. 2016 Feb 29;7:210. doi: 10.3389/fmicb.2016.00210. eCollection 2016. Front Microbiol. 2016. PMID: 26973602 Free PMC article.
-
Protocells and RNA Self-Replication.Cold Spring Harb Perspect Biol. 2018 Sep 4;10(9):a034801. doi: 10.1101/cshperspect.a034801. Cold Spring Harb Perspect Biol. 2018. PMID: 30181195 Free PMC article. Review.
References
-
- Allison J. D., Brown D. S., Novo-Gradac K. J. (1991). MINTEQ A2/PRODEFA2, a Geochemical Assessment Model for Environmental Systems: Version 3.0 User's Manual. Athens: Environmental Research Laboratory, Office of Research and Development, USEPA.
-
- APHA (1998). Standard Methods for the Examination of Water and Wastewater. Washington, DC: American Public Health Association.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

